Researchers using the Stanford University’s Linac Coherent Light Source – the world’s first hard X-ray free-electron laser and one of only two currently operating in the world – have produced a 3D image revealing the inner structure of a Mimivirus virion.
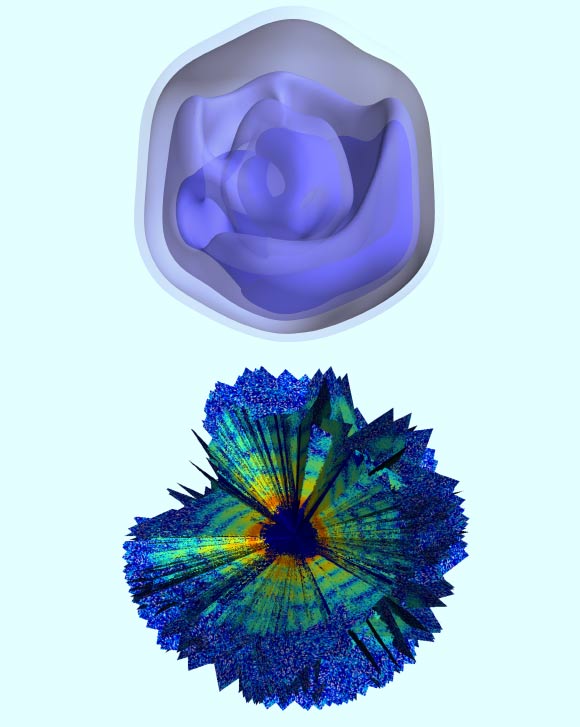
Top: 3D reconstruction of a Mimivirus virion; the blue regions represent the areas of highest density. Bottom: computerized rendering shows a cutaway view of a collection of about 200 X-ray patterns; the images were combined to produce a 3D rendering of the Mimivirus particle. Image credit: Uppsala University.
First isolated in 1992 from Acanthamoeba polyphaga growing in a water tower in Bradford, UK, Mimivirus is one of the largest, most complex viruses known.
The Mimivirus’ structure is so similar to that of the amoeba that it was only identified as a virus in 2003.
The virus appears to be an icosahedral-shaped particle with a diameter of 400 nm and no envelope, surrounded by 80-nm-long fibrils.
It has a double-stranded DNA circular genome of about 800 kilobase pairs.
Both the genome and the virion size of the virus are larger than that of some small bacteria.
Researchers have been trying to determine the inner structure of this and other giant viruses to learn more about their origins.
For example, did they borrow genes over time from the host organisms they infect, like amoebas? Did they precede cell-based life or devolve from cell-based organisms?
In the new study, scientists led by Dr Tomas Ekeberg of Uppsala University in Sweden sprayed a gas-propelled aerosol containing active Mimivirus virions in a thin stream into the X-ray laser beam, which scattered off the virions and produced light patterns on a detector that were recorded as diffraction images.
They then compiled hundreds of individual images from separate virions into a single 3D portrait showing the general shape and inner features of Mimivirus.
“We can see quite clearly that the inside of these viruses is not uniform,” said Dr Ekeberg, who is the lead author of the paper published in the journal Physical Review Letters.
The results pose an important step towards solving the structures of non-crystallizable biomolecules, demonstrating that it’s possible to gather noisy diffraction data from hundreds of individual samples and assemble the data into a complete diffraction pattern, from which the 3D structure can be derived.
The technique’s spatial resolution – currently 125 nm – will need to be improved before biomolecules like proteins, much smaller than the giant Mimivirus, can be tackled.
However, the method could already image important pathogenic viruses like HIV, influenza and herpes.
“The research team now plans to apply the 3D imaging technique to other types of samples and to improve the image quality. The next Holy Grail is to study large, single proteins,” said Prof Janos Hajdu of Uppsala University, the senior author of the paper.
_____
Tomas Ekeberg et al. 2015. Three-Dimensional Reconstruction of the Giant Mimivirus Particle with an X-Ray Free-Electron Laser. Phys. Rev. Lett. 114, 098102; doi: 10.1103/PhysRevLett.114.098102







