Researchers from Austria and the United States have identified a group of giant viruses — Klosneuviruses — that harbor components of many other viruses and proteins, and their analyses suggest that Klosneuviruses acquired the various components in an evolutionarily recent time frame, likely from, and as an adaptation to, their hosts. A paper describing this discovery was published in the April 7 issue of the journal Science.
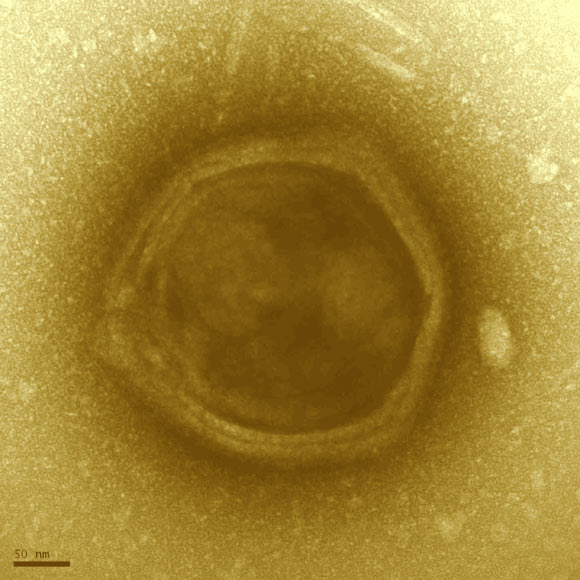
Transmission electron microscopy image of a candidate Klosneuvirus particle detected in the Klosterneuburg waste water treatment plant biomass. The particle exhibits features reminiscent of Mimiviridae including a faceted, multi-layered envelope surrounding a central core. Scale bar – 50 nm. Image credit: Schulz et al.
Biologists have been fascinated by giant viruses since 2003, when a research team led by CNRS scientist Didier Raoult discovered and characterized Mimiviruses.
Since then, a handful of other giant virus groups, such as Pandoraviruses and Pithoviruses, have been found.
The unique ability among them to encode proteins involved in translation piqued researchers’ interests as to the origin of giant viruses.
Since then, two evolutionary hypotheses have emerged:
(i) one posits that giant viruses evolved from an ancient cell, perhaps one from an extinct fourth domain of cellular life;
(ii) another presents the idea that giant viruses arose from smaller viruses.
“The discovery of Klosneuvirus supports the latter idea,” said lead co-author Dr. Tanja Woyke, Microbial Genomics Program lead at the U.S. Department of Energy Joint Genome Institute.
“In this scenario, a smaller virus infected different eukaryote hosts and picked up genes encoding translational machinery components from independent sources over long periods of time through piecemeal acquisition.”
Dr. Woyke and co-authors found Klosneuvirus in the wastewater of a treatment plant in Austria.
“Analysis of low-complexity metagenomes from a wastewater treatment plant in Klosterneuburg, Austria, revealed clearly separated genomic bins comprising many genes typically found in giant viruses,” they explained.
“From these data, a 1.57-Mb genome of a putative virus, which we named Klosneuvirus, was assembled.”
To detect viruses related to Klosneuvirus, the team screened nearly 7,000 environmental metagenomes and discovered “three metagenomics bins with high assembly quality and strong overlap in gene content.”
“All three bins were identified as giant virus genomes, ranging from 0.86 Mb (Indivirus) to 1.33 Mb (Hokovirus) to 1.53 Mb (Catovirus),” the scientists said.
At first glance, the suite of ‘cellular’ genes in Klosneuviruses seemed to have a common origin, but when analyzing them in detail, the authors observed they came from different hosts.
From the evolutionary trees the scientists built, they noticed that they were acquired by the viruses bit by bit, at different stages in their evolution.
The Klosneuvirus genes contained aminoacyl-tRNA (transfer ribonucleic acid) enzymes with specificity for 19 out of 20 amino acids, along with more than 20 tRNAs and an array of translation factors and tRNA modifying enzymes — an unprecedented finding among all viruses, including the previously known giant viruses.
The predicted hosts for Klosneuviruses are protists and while their direct impacts on protists are not yet worked out, these giant viruses are thought to have a large impact on these protists that help regulate the planet’s biogeochemical cycles.
The team also determined that Klosneuviruses belong to a family called Mimiviridae.
“The discovery presents virus evolution for us in new ways, vastly expanding our understanding of how many essential host genes viruses can capture during their evolution,” said co-author Dr. Eugene Koonin, an evolutionary and computational biologist at the National Center for Biotechnology Information at the National Institutes of Health.
“Since protein synthesis is one of the most prominent hallmarks of cellular life, it shows that these new viruses are more ‘cell-like’ than any virus anyone has ever seen before.”
_____
Frederik Schulz et al. 2017. Giant viruses with an expanded complement of translation system components. Science 356 (6333): 82-85; doi: 10.1126/science.aal4657







