A set of three nearly identical genes found only in humans, NOTCH2NL, appears to play a critical role in the development of our large brains, according to a pair of studies published this week in the journal Cell. The NOTCH2NL genes are found exclusively in humans and appeared between 3 and 4 million years ago, just before the period when fossils show a dramatic increase in the brain sizes of human ancestors. These genes belong to an ancient family of genes known as NOTCH genes, first discovered in fruit flies and named for a genetic defect causing notched wings.
The NOTCH2NL genes arose in human ancestors just before a dramatic increase in brain size and are involved in genetic defects associated with neurological disorders. Image credit: Gerd Altmann / Sci-News.com.
The evolution of larger brains in the last 3 million years played an important role in our ability as a species to think, problem-solve, and develop culture. But the genetic changes behind the expansion that made us human have been elusive.
“Our brains got three times as big primarily through the expansion of certain functional areas of the cerebral cortex, and that has to be a fundamental substrate for us becoming human,” said Dr. David Haussler, co-lead author of one of the Cell papers and a researcher at the University of California, Santa Cruz, and Howard Hughes Medical Institute.
“There’s really no more exciting scientific question that I can think of than discovering and decoding the mysterious genetic changes that made us who we are.”
The newly-identified human-specific genes were derived from NOTCH2, one of four mammalian NOTCH genes, through a duplication event that inserted an extra partial copy of NOTCH2 into the genome.
This happened in an ancient ape species that was a common ancestor of humans, chimpanzees, and gorillas. The partial duplicate was a nonfunctional ‘pseudogene,’ versions of which are still found in chimp and gorilla genomes.
In the human lineage, however, this pseudogene was ‘revived’ when additional NOTCH2 DNA was copied into its place, creating a functional gene. This new gene was then duplicated several more times, resulting in four related NOTCH2NL genes.
One of the four NOTCH2NL genes appears to be a nonfunctional pseudogene, but the other three (NOTCH2NLA, NOTCH2NLB, and NOTCH2NLC) are active genes that direct the production of truncated versions of the original NOTCH2 protein.
“We were comparing genes expressed during brain development in humans and macaque monkeys in stem cell-derived models when we realized that we could detect NOTCH2NL in human cells but not in those of the macaques,” the researchers explained.
“Looking at the DNA, we also didn’t see it in orangutans and found only truncated, inactive versions in our closest relatives, gorillas and chimpanzees.”
“Reconstructing the evolutionary history of NOTCH2NL genes revealed that a process called gene conversion was likely responsible for repairing a non-functional version of NOTCH2NL, which originally emerged as a partial duplication of an essential neurodevelopmental gene known as NOTCH2.”
“This repair happened only in humans — and we estimate it happened 3-4 million years ago, around the same time that the fossil record suggests human brains began to expand.”
“After it was repaired, but before we diverged from our common ancestor with Neanderthals, NOTCH2NL was duplicated two more times.”
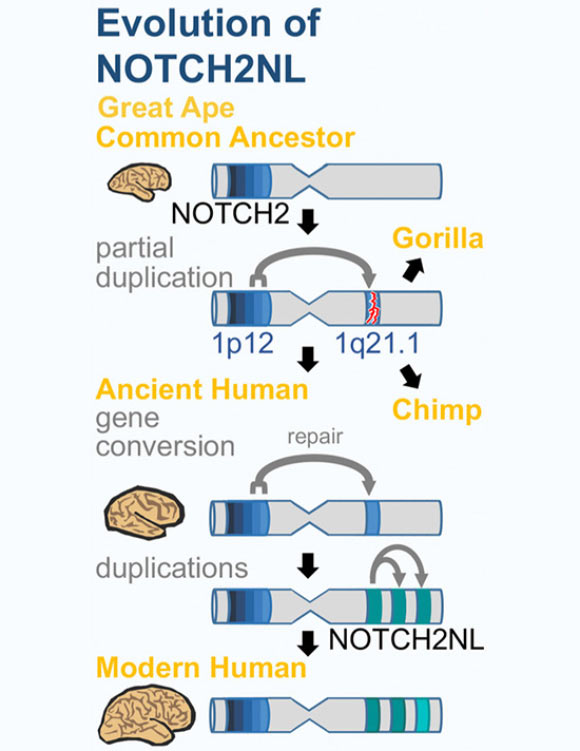
As shown in this diagram, the NOTCH2NL genes arose through a duplication event in a common ancestor of humans, chimpanzees, and gorillas, but only became functional genes in the human lineage. Image credit: Sofie Salama.
The team behind the other Cell paper, led by developmental biologist Pierre Vanderhaeghen of Université Libre de Bruxelles and VIB-KU Leuven, arrived at NOTCH2NL from a related direction, searching for human-specific genes active during fetal brain development using primary tissue.
“One of the holy grails of researchers like us is to find out what during human development and evolution is responsible for a bigger brain, particularly the cerebral cortex. Given the relatively fast evolution of the human brain, it is tempting to speculate that newly evolved, human-specific genes may help shape our brain in a species-specific way,” Dr. Vanderhaeghen said.
Searching for human-specific genes involved in brain development proved challenging because these genes are typically poorly annotated in genome databases and hard to distinguish from more common genes present in other species.
Dr. Vanderhaeghen’s team developed a tailored RNA sequencing analysis for specific and sensitive detection of human-specific genes in human fetal cerebral cortex.
This allowed the study authors to identify a repertoire of 35 genes unique to humans that are active during development of the cerebral cortex in humans, including NOTCH2NL genes.
They zeroed in on NOTCH2NL in particular because of the importance of its ancestral gene, NOTCH2, in signaling processes that control whether cortical stem cells produce neurons or regenerate more stem cells. And they found that artificially expressing NOTCH2NL in mouse embryos increased the number of progenitor stem cells in the mouse cortex.
To better understand what the genes do in humans, the scientists turned to an in vitro model of cortical development from human pluripotent stem cells to explore NOTCH2NL function.
In this model, they found that NOTCH2NL can substantially expand the population of cortical stem cells, which in turn then generate more neurons, a feature expected to distinguish between human and non-human cortical neurogenesis.
“From one stem cell, you can either regenerate two progenitor cells, generate two neurons, or generate one progenitor stem cell and one neuron,” Dr. Vanderhaeghen said.
“And what NOTCH2NL does is bias that decision in a slight way towards regenerating progenitors, which can later go on to make more neurons. It’s a small early effect with large late consequences, as often happens with evolution.”
Dr. Haussler and colleagues looked at what happened when NOTCH2NL wasn’t expressed: they deleted it from human stem cells and used them to grow patches of cortex called organoids.
In the organoids derived from NOTCH2NL-depleted stem cells, differentiation occurred faster, but the organoids ended up being smaller.
“If you lose NOTCH2NL, it leads to premature differentiation of cortical stem cells into neurons, but at the same time the very important stem cell pool gets depleted,” said University of Amsterdam researcher Dr. Frank Jacobs, a member of Dr. Haussler’s team.
NOTCH2NL’s location on the genome, incorrectly mapped until recently, is further support for its role in human brain size.
Duplications or deletions at a genome region known as 1q21.1 are known to cause macrocephaly or microcephaly, respectively, and are associated with a range of neurodevelopmental disorders, including ADHD, autism spectrum disorder, and intellectual disability.
Dr. Haussler’s team looked at 11 patients with errors at this locus and found that NOTCH2NL was indeed being duplicated and deleted in the rearrangement events associated with larger and smaller brain size that resulted.
“We really wanted the gene to be in the 1q21.1 disease interval, because it made logical sense, but in the incorrect reference genome, it wasn’t. And then we found new data, and we realized that it was a mistake in the reference genome! It seldom happens that when you want something that appears to be false to be true, it turns out to actually be true. I don’t think something of that level will ever happen again in my career,” Dr. Haussler said.
Because NOTCH2NL is something of an evolutionary trade-off between larger brains and 1q21.1 disease susceptibility, the researchers are all quick to point out that there is plenty of healthy variation here, too.
“It’s a boon that may have enabled us to get a big brain. And yes, it’s a bane, because we can have these recombination events that can be bad. But what we found when we developed the technology to really sequence it in individuals is that there are multiple different alleles of this gene. And it’s possible that that variation creates the subtlety and plasticity that is important in enabling humans to be human,” said Dr. Sofie Salama, a researcher at the University of California, Santa Cruz, and Howard Hughes Medical Institute, and a member of Dr. Haussler’s team.
_____
Ian T. Fiddes et al. 2018. Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell 173 (6): 1356-1369; doi: 10.1016/j.cell.2018.03.051
Ikuo K. Suzuki et al. 2018. Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell 173 (6): 1370-1384; doi: 10.1016/j.cell.2018.03.067







