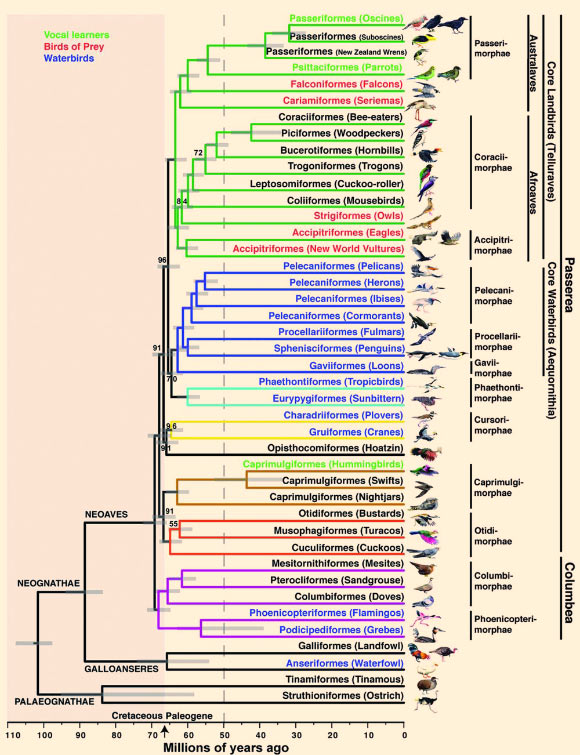
In an ambitious four-year genetic study on bird evolution, an international consortium of more than 200 scientists from 80 institutions across 20 countries has sequenced, assembled and compared the full genomes of 48 bird species representing all major branches of modern birds – including the woodpecker, owl, penguin, hummingbird and flamingo.
The scientists are publishing their findings nearly simultaneously this week in 28 papers – eight papers in a special issue of the journal Science and 20 more in Genome Biology, BMC Genomics, GigaScience and other journals.
They compared the 48 bird genomes they sequenced with those of three other reptile species and humans. This enabled them to investigate at which point in each species’ history specialized characteristics developed, such as feathers, flight and song.
One of the Science papers found there is a consistent set of just over 50 genes that show higher or lower activity in the brains of vocal learning birds and humans. These changes were not found in the brains of birds that do not have vocal learning and of non-human primates that do not speak.
“This means that vocal learning birds and humans are more similar to each other for these genes in song and speech brain areas than other birds and primates are to them,” said Dr Erich Jarvis of Duke University Medical Center, the senior author of the paper.
These genes are involved in forming new connections between neurons of the motor cortex and neurons that control the muscles that produce sound.
The scientists also looked at the specialized activity of a pair of genes involved in the regions of the brain that control song and speech. They found that these genes are down- and up-regulated in one brain region of song-learning birds during the juvenile period of their vocal learning, changes that last into adulthood. They hypothesize that changes in these genes could be critical for the evolution of song in birds and speech in humans. The results appear in the Journal of Comparative Neurology.
Among the three main groups of vocal learning birds, parrots are clearly different in their ability to mimic human speech. The scientists used the activity of some of the specialized genes to discover that the parrot’s speech center is organized somewhat differently. It has what they call a ‘song-system-within-a-song-system’ in which the area of the brain with different gene activity for producing song has an outer ring of still more differences in gene expression.
“Parrots are very social animals and having the ability to quickly pick up dialects of parrot speech may account for their super-charged speech center. The shell or outer regions were found to be proportionally larger in the parrot species, which are believed to have the highest vocal, cognitive and social abilities. These species include Amazon parrots, the African Grey and the Blue and Gold Macaw,” said Dr Mukta Chakraborty of Duke University Medical Center, who is a co-author of the Journal of Comparative Neurology paper.
The team looked at gene activation in different areas of the brain during singing. They found activation of 10 percent of the expressed genome during singing, with diverse activation patterns in different song-learning regions of the brain. The diverse gene patterns are best explained by epigenetic differences in the genomes of the different brain regions, meaning that individual cells in different brain regions can regulate genes at a moment’s notice when the birds sing.
The scientists also found the genomes of vocal learners are more rapidly evolving and have more chromosomal rearrangements compared to other bird species. This genomic comparison also found similar changes occurred independently in the song-learning area of different birds’ brains.
“Speech is difficult to study in human brains. Whales and elephants learn speech and songs, but they’re too big to house in the lab. Now that we have a deeper understanding of how similar birdsong brain regions are to human speech regions at the genetic level, I think they’ll be a better model than ever,” Dr Jarvis said.
The researchers also sequenced three crocodilian genomes – the American alligator’s, the saltwater crocodile’s, and the Indian gharial’s – which represent birds’ closest living relatives. The genomes of such crocodilians are evolving at an exceptionally slow pace.
“The molecular evolution of birds is much faster than it is in crocs, turtles and other reptilian lineages. So this avian lineage seems to be faster than other reptiles, but not faster than mammals,” said lead author Dr Richard Green from the University of California, Santa Cruz.
A separate report suggests that the mutations that eliminated enamel and dentin from the teeth of modern birds – an event which eventually led to toothless beaks – began about 116 million years ago.
Another study in Science describes how to produce the most accurate phylogenetic trees from gene trees with a technique they call ‘statistical binning.’
Do bird genomes carry fewer virus sequences than other species? Mammalian genomes harbor a diverse set of genomic ‘fossil’s of past viral infections called endogenous viral elements (EVE). A paper published in the journal Genome Biology found that bird species had 6-13 times fewer EVE infections in their past than mammals. This finding is consistent with the fact that birds have smaller genomes than mammals. It also suggests birds may either be less susceptible to viral invasions or better able to purge viral genes.
_____
Qi Zhou et al. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science, vol. 346, no. 6215; doi: 10.1126/science.1246338
Jie Cui et al. 2014. Low Frequency of Paleoviral Infiltration Across the Avian Phylogeny. Genome Biology, 15:539; doi: 10.1186/s13059-014-0539-3
Robert W. Meredith et al. 2014. Evidence for a single loss of mineralized teeth in the common avian ancestor. Science, vol. 346, no. 6215; doi: 10.1126/science.1254390
Osceola Whitney et al. 2014. Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science, vol. 346, no. 6215; doi: 10.1126/science.1256780
Andreas R. Pfenning et al. 2014. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science, vol. 346, no. 6215; doi: 10.1126/science.1256846
Rui Wang et al. Convergent differential regulation of SLIT-ROBO axon guidance genes in the brains of vocal learners. Journal of Comparative Neurology, published online November 26, 2014; doi: 10.1002/cne.23719
Siavash Mirarab et al. 2014. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science, vol. 346, no. 6215; doi: 10.1126/science.1250463
Richard E. Green et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science, vol. 346, no. 6215; doi: 10.1126/science.1254449