Researchers have made a breakthrough toward developing a vaccine for the SARS-CoV-2 coronavirus, also known as 2019-nCoV, by creating the first 3D atomic scale map of its spike protein, part of a virus that attaches to and infects human cells.
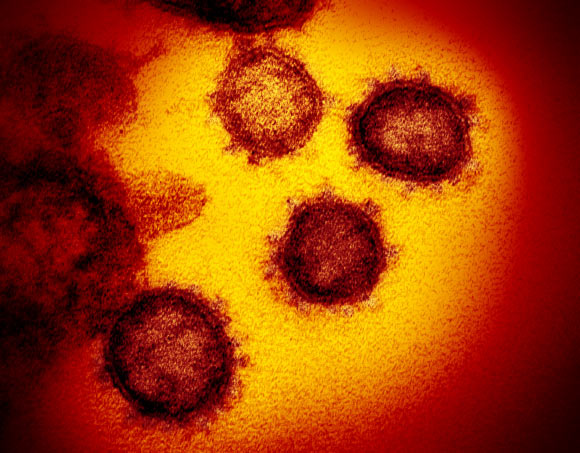
This transmission electron microscope image shows SARS-CoV-2, also known as 2019-nCoV, the virus that causes COVID-19, isolated from a patient in the U.S., emerging from the surface of cells cultured in the lab. Image credit: NIAID-RML.
“The novel coronavirus has recently emerged as a human pathogen in the city of Wuhan in China’s Hubei province, causing fever, severe respiratory illness and pneumonia, a disease recently named COVID-19,” said senior author Dr. Jason McLellan from the Department of Molecular Biosciences at the University of Texas at Austin and colleagues.
“The emerging pathogen was rapidly characterized as a novel member of the betacoronavirus genus, closely related to several bat coronaviruses as well as SARS-CoV.”
“Compared to SARS-CoV, SARS-CoV-2 appears to be more readily transmitted from human-to-human, spreading to multiple continents.”
“It makes use of a densely glycosylated spike protein to gain entry into host cells.”
Dr. McLellan team had already developed methods for locking spike proteins in other coronaviruses, including SARS-CoV and MERS-CoV, into a shape that made them easier to analyze and could effectively turn them into candidates for vaccines.
This experience gave them an advantage over other research teams studying the novel virus.
“As soon as we knew this was a coronavirus, we felt we had to jump at it, because we could be one of the first ones to get this structure,” Dr. McLellan said.
“We knew exactly what mutations to put into this, because we’ve already shown these mutations work for a bunch of other coronaviruses.”
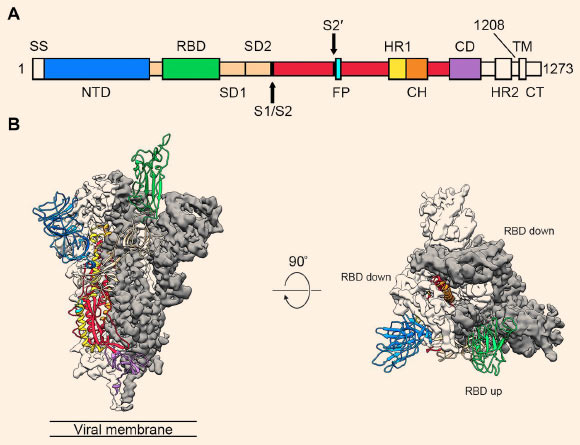
Structure of SARS-CoV-2’s spike protein: (A) schematic of the spike protein’s primary structure, colored by domain; domains that were excluded from the ectodomain expression construct or could not be visualized in the final map are colored white. Abbreviations: SS – signal sequence, NTD – N-terminal domain, RBD – receptor-binding domain, SD1 – subdomain 1, SD2 – subdomain 2, S1/S2 – S1/S2 protease cleavage site, S2′ – S2′ protease cleavage site, FP – fusion peptide, HR1 – heptad repeat 1, CH – central helix, CD – connector domain, HR2 – heptad repeat 2, TM – transmembrane domain, CT – cytoplasmic tail; arrows denote protease cleavage sites; (B) side and top views of the prefusion structure of SARS-CoV-2’s spike protein with a single RBD in the up conformation; the two RBD-down protomers are shown as cryo-EM density in either white or gray and the RBD-up protomer is shown in ribbons, colored corresponding to the schematic in (A). Image credit: Wrapp et al, doi: 10.1126/science.abb2507.
Just two weeks after receiving the genome sequence of the SARS-CoV-2 virus, the researchers had designed and produced samples of its stabilized spike protein.
It took about 12 more days to reconstruct the 3D atomic scale map, called a molecular structure, of the spike protein.
Critical to the success was state-of-the-art technology known as cryogenic electron microscopy (cryo-EM), which allows scientists to make atomic-scale 3D models of cellular structures, molecules and viruses.
The molecule the team produced, and for which they obtained a structure, represents only the extracellular portion of the spike protein, but it is enough to elicit an immune response in people, and thus serve as a vaccine.
Next, the authors plan to use their molecule to pursue another line of attack against the SARS-CoV-2 virus, using the molecule as a ‘probe’ to isolate naturally produced antibodies from patients who have been infected with the novel coronavirus and successfully recovered.
In large enough quantities, these antibodies could help treat a coronavirus infection soon after exposure. For example, the antibodies could protect soldiers or health care workers sent into an area with high infection rates on too short notice for the immunity from a vaccine to take effect.
The team’s results are published in the journal Science.
_____
Daniel Wrapp et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, published online February 19, 2020; doi: 10.1126/science.abb2507







