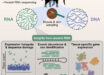
An international team of scientists has obtained and analyzed the genome of five barley grains that date back to the Chalcolithic period, about 6,000 years ago.

Left: 6,000-year-old plant remains in Yoram Cave; note the excellent dry preservation of reeds, seeds and pellets. Right: well-preserved desiccated barley grain found at Yoram Cave. Scale bar – 2 mm. Image credit: Martin Mascher et al.
The 6,000-year-old barley grains were unearthed in Yoram Cave, part of a complex of three difficult-to-access caves in the Judean Desert, Israel.
“Yoram Cave is archaeologically significant as one of the rare cave sites with a single layer of human occupation according to current radiocarbon dating findings,” the authors said.
“Unlike most other Judean Desert caves, there are no findings from the later Roman and Byzantine Periods. In addition, it is one of the rare cave sites that have not suffered from modern looting or hyena burrowing.”
“It is the only Chalcolithic cave site in the Judean Desert that has been excavated by high-resolution sampling methods.”
The genome of the Chalcolithic barley grains is the oldest plant genome reconstructed to date, according to the researchers who published their findings in the journal Nature Genetics.
“These archaeological remains provided a unique opportunity for us to finally sequence a Chalcolithic plant genome,” said senior co-author Dr. Ehud Weiss, from Bar-Ilan University.
“The genetic material has been well-preserved for several millennia due to the extreme dryness of the region.”
In order to determine the age of the barley grains from Yoram Cave, the scientists split the grains and subjected half of them to radiocarbon dating while the other half was used to extract the ancient DNA.
They also compared the ancient barley genome sequences to the genomes of extant wild (Hordeum vulgare ssp. spontaneum) and domesticated (Hordeum vulgare ssp. vulgare) barley.
“The comparison of the ancient seeds with wild forms from the region and barley landraces (local barley lines grown by farmers in the Near East) allowed the team to suggest the origin of the domestication of barley within the Upper Jordan Valley – a hypothesis that is also supported by two archaeological sites in the surrounding area where the hitherto earliest remains of barley cultivation have been found,” said senior co-author Dr. Tzion Fahima, from the University of Haifa.
The team’s findings suggest that barley landraces grown in present-day Israel have not experienced major lineage turnover over the past 6,000 years, although there is evidence for gene flow between cultivated and wild populations.
“Despite hybridization events between wild and domesticated barleys over the last six millennia, the overall picture is that the genomes of extant Levantine landraces have remained remarkably similar to how they were 6,000 years ago,” the scientists explained.
“This is despite climate change and anthropogenic transformations of local flora and fauna, including changes in agricultural practices, which might have favored the introduction of landraces from other regions that were better adapted to the changing agricultural environment.”
“Although we found no indications of major lineage turnovers in the barley crop from the Southern Levant, the eventful history of this region makes it likely that the farmers who grew cereals there several millennia ago are not the ancestors of those who tend the present-day landraces. One can speculate that conquerors and immigrants did not bring crop seeds from their old homelands but favored locally adapted landraces.”
_____
Martin Mascher et al. Genomic analysis of 6,000-year-old cultivated grain illuminates the domestication history of barley. Nature Genetics, published online July 18, 2016; doi: 10.1038/ng.3611