Oat (Avena sativa) grain is a traditional human food that is rich in dietary fiber and contributes to improved human health. Interest in the crop has surged in recent years owing to its use as the basis for plant-based milk analogues. In contrast to many other cereal species, genomic research in oat is still at an early stage, and surveys of structural genome diversity and gene expression variability are scarce. Scientists have now assembled and annotated the genomes of 33 wild and domesticated oat lines, along with an atlas of gene expression across 6 tissues of different developmental stages in 23 of these lines.
Oat — the world’s seventh most widely grown cereal crop — is appreciated for its high content of dietary fiber, which has been shown to have substantial benefits for human health.
In 2022 and 2023, more than 25 million metric tons were produced worldwide.
Genetically improved cultivars have the potential to make oat cultivation more productive and sustainable, but much of this potential remains unrealized, and the first oat reference sequences have been published only in the past few years.
The complexity of the oat genome is partly to blame for the slow progress.
“The pangenome is central to our understanding of cultivated plants such as oats, as it maps their entire genetic diversity,” said first author Dr. Raz Avni, a researcher at the Leibniz Institute of Plant Genetics and Crop Plant Research, and colleagues.
“It encompasses not only genes that occur in all plants, but also those that are only present in certain species, serving as a kind of map.”
“In turn, the pantranscriptome shows which genes are active in different tissues, such as leaves, roots and seeds, and at different stages of development. It serves as a gene expression atlas.”
“However, understanding how genetic differences influence individual plant traits is challenging, particularly in the case of oats.”
“The oat genome is very complex because oats are a hexaploid plant with six sets of chromosomes originating from three different ancestors.”
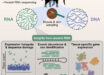
In the research, the authors sequenced and analyzed the genomes of 33 oat lines, encompassing cultivated varieties and their wild relatives.
They also created the oat pantranscriptome by examining the gene expression patterns in six tissues and developmental stages of 23 of these oat lines.
The aim was to identify possible structural variations. These can involve changes in the arrangement of chromosomes, such as inversions (i.e. sections that have been rotated) or translocations (i.e. sections that have been moved to a different location).
“With our pangenome, we demonstrate the true extent of the genetic diversity in oats,” Dr. Avni said.
“This helps us to better understand which genes are important for yield, adaptation, and health.”
The researchers also came across some surprising details in their work.
“For instance, we found that many genes had been lost in one of the three subgenomes,” they said.
“However, the plant remains productive because other gene copies apparently take over the corresponding tasks.”
“Decoding the oat pangenome shows how modern genomics can advance basic research and have a direct impact on health, agriculture, and breeding,” said senior author Dr. Martin Mascher, a researcher at the Leibniz Institute of Plant Genetics and Crop Plant Research, Murdoch University and the German Centre for Integrative Biodiversity Research.
“We have also found that structural variation in the genome affects genes responsible for controlling flowering time.”
The team’s results appear in the journal Nature.
_____
R. Avni et al. A pangenome and pantranscriptome of hexaploid oat. Nature, published online October 29, 2025; doi: 10.1038/s41586-025-09676-7