Modern individuals from the Pacific islands of Melanesia harbor adaptive copy number variants (CNVs) that they inherited from two groups of our evolutionary cousins, Neanderthals and Denisovans, according to new research led by the University of Washington School of Medicine.

A portrait of a juvenile female Denisovan based on a skeletal profile reconstructed from ancient DNA methylation maps. Image credit: Maayan Harel.
Genetic surveys with single-nucleotide variants (SNVs) have suggested their involvement in archaic introgression and adaptation.
However, CNVs — a larger form of structural variant — are far more likely to be associated with genotype expression and are subject to stronger selective pressure.
Despite this, the adaptive role of introgressed CNVs in human evolution and the genetic variation of modern humans remains unexplored.
Dr. PingHsun Hsieh from the Department of Genome Sciences at the University of Washington School of Medicine and his colleagues performed a genome-wide search for evidence of selective and archaic introgressed CNVs among the genomes of Melanesians, an Oceanian population distributed from Papua New Guinea to as far east as the islands of Fiji and known to harbor some of the greatest amounts of Neanderthal and Denisovan ancestry.
The researchers examined 266 modern human genomes from the Simons Genome Diversity Project and the genomes of three ancient hominins: a Denisovan, a Neanderthal from the Altai Mountains in Siberia, and a Neanderthal from Croatia.
They discovered hominin-shared, stratified CNVs associated with positive selection in the modern Melanesian genomes.
Furthermore, the results revealed evidence for adaptive CNVs introgression at chromosomes 16p11.2 and 8p21.3, which were derived from Denisovans and Neanderthals, respectively, and are absent from most other human populations.
At chromosome 16p11.2, the scientists found a large duplication of over 383,000 base pairs that originated from Denisovans and introgressed into the ancestral Melanesian population 60,000 to 170,000 years ago.
On chromosome 8p21.3, they identified a Melanesian haplotype that carries two CNVs with a Neanderthal origin and that introgressed into non-Africans 40,000 to 120,000 years ago.
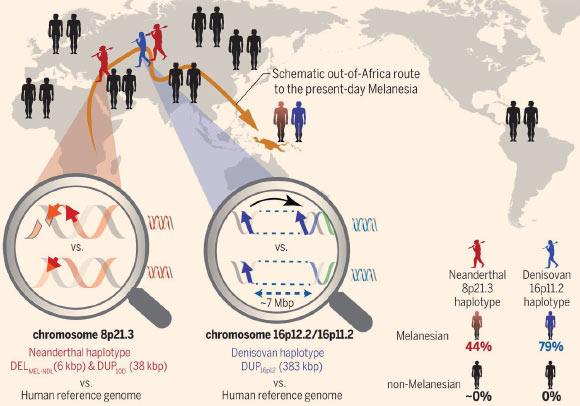
Large adaptive-introgressed CNVs at chromosomes 8p21.3 and 16p11.2 in Melanesians. The magnifying glasses highlight structural differences between the archaic (top) and reference (bottom) genomes. Neanderthal (red) and Denisovan (blue) haplotypes encompassing large CNVs occur at high frequencies in Melanesians (44 and 79%, respectively) but are absent (black) in all non-Melanesians. These CNVs create positively selected genes (TNFRSF10D1, TNFRSF10D2, and NPIPB16) that are absent from the reference genome. Image credit: Hsieh et al, doi: 10.1126/science.aax2083.
“Our results suggest that large structural variants introgressed from our archaic ancestors may have played an important role in the adaptation of early modern human populations and that they may represent an under-appreciated source of the genetic variation that remains to be characterized in our modern genomes,” the study authors said.
“As populations of our ancestors migrated out of Africa and into the vast Eurasian continent, they were required to adapt to the wide range of environments they encountered. They also interbred with the archaic hominin ancestors they encountered.”
“However, the role of genetic exchange between archaic hominin and anatomically modern human populations in adaptation and human evolution remains elusive.”
The results were published in the October 18, 2019 issue of the journal Science.
_____
PingHsun Hsieh et al. 2019. Adaptive archaic introgression of copy number variants and the discovery of previously unknown human genes. Science 366 (6463): eaax2083; doi: 10.1126/science.aax2083







