The COVID-19 pandemic caused by the SARS-CoV-2 coronavirus is a global health emergency. A team of researchers in Germany has figured out the structure of SARS-CoV-2’s main protease, a drug target among coronaviruses.
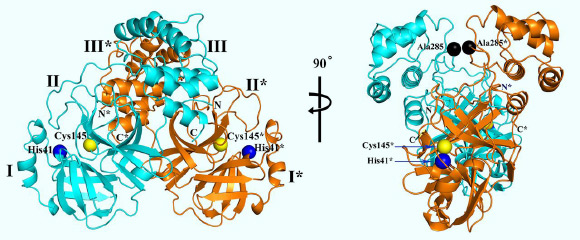
3D structure of SARS-CoV-2 Mpro, in two different views. One protomer of the dimer is shown in light blue, the other one in orange. Domains are labeled by Roman numbers. Amino-acid residues of the catalytic site are indicated as yellow and blue spheres, for Cys145 and His41, respectively. An asterisk marks a residue from protomer B (orange). Black spheres indicate the positions of Ala285 of each of the two domains III. Chain termini are labeled N and C for molecule A (light blue) and N* and C* for molecule B (orange). Image credit: Zhang et al, doi: 10.1126/science.abb3405.
In December 2019, a new coronavirus caused an outbreak of pulmonary disease in the city of Wuhan, the capital of Hubei province in China, and has since spread globally.
The virus has been named SARS-CoV-2, because the RNA genome is about 82% identical to the SARS-CoV, a coronavirus that causes SARS.
The disease caused by SARS-CoV-2 is called COVID-19. Whereas at the beginning of the outbreak, cases were connected to the Huanan seafood and animal market in Wuhan, efficient human-to-human transmission led to exponential growth in the number of cases.
On March 11, the World Health Organization declared the outbreak a pandemic.
One of the best characterized drug targets among coronaviruses is the main protease: Mpro, also called 3CLpro.
“Teams around the world are working hard to develop active substances against SARS-CoV-2. The structural analysis of functional proteins of the virus is very helpful for this goal,” said University of Lübeck’s Professor Rolf Hilgenfeld and colleagues.
“The function of a protein is closely related to its 3D architecture. If this 3D architecture is known, it is possible to identify specific points of attack for active substances.”
The researchers decoded the 3D structure of the SARS-CoV-2 Mpro using the high-intensity X-ray light from the BESSY II facility at the Helmholtz-Zentrum Berlin.
“For such issues of highest relevance, we can offer fast track access to our instruments,” said Dr. Manfred Weiss, head of the Research Group Macromolecular Crystallography at the Helmholtz-Zentrum Berlin.
“Tiny protein crystals can be analyzed with highly brilliant X-ray light,” the scientists explained.
“The images contain information about the 3D architecture of the protein molecules.”
“The complex shape of the protein molecule and its electron density is then calculated by computer algorithms.”
“The 3D architecture provides concrete starting points for developing active substances or inhibitors. These drugs could dock specifically to target points of the macromolecule and impede its function.”
The results are published in the journal Science.
_____
Linlin Zhang et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, published online March 20, 2020; doi: 10.1126/science.abb3405