In a study that has implications to advance medicine and biodiversity conservation, a large international consortium of researchers involved in the Zoonomia Project has sequenced and analyzed the genomes of 131 species of placental mammals, bringing the worldwide total to 240.

The Zoonomia Project brings the fraction of eutherian mammal families that are represented by at least one assembly to 83%. This image shows the brown-throated sloth (Bradypus variegatus) in the Cahuita National Park, Costa Rica. Image credit: Christian Mehlführer / CC BY 2.5.
The genomics revolution is enabling advances not only in medical research, but also in basic biology and in the conservation of biodiversity, where genomic tools have helped to apprehend poachers and to protect endangered populations.
However, we have only a limited ability to predict which genomic variants lead to changes in organism-level phenotypes, such as increased disease risk — a task that, in humans, is complicated by the sheer size of the genome.
Comparative genomics can address this challenge by identifying nucleotide positions that have remained unchanged across millions of years of evolution, focusing the search for disease-causing variants.
In 2011, the 29 Mammals Project identified genetic regions of evolutionary constraint that in total comprise 4.2% of the genome, by measuring sequence conservation in humans plus 28 other mammals.
These regions proved to be more enriched for the heritability of complex diseases than any other functional mark, including coding status.
By expanding the number of species and making an alignment that is independent of any single reference genome, the Zoonomia Project — formerly called the 200 Mammals Project — was designed to detect evolutionary constraint in the eutherian lineage at increased resolution, and to provide genomic resources for over 130 previously uncharacterized species.
“The comparison of the genomes from the 240 mammals will help geneticists to identify the mutations that lead to human diseases,” said Professor Kerstin Lindblad-Toh, a researcher at Uppsala University, SciLifeLab and the Broad Institute of MIT and Harvard.
The Zoonomia scientists identified genetic innovations that seem to protect certain animals from diseases like cancer and diabetes.
They also pinpointed genomic elements that have remained unchanged across millions of years of evolution, which predict where mutations are likely to be associated with risk of disease, and could reveal new avenues of therapeutic development.
“Before the microscope, we couldn’t see what was going on inside of a cell,” said Dr. Oliver Ryder, Kleberg endowed director of conservation genetics at the San Diego Zoo Institute for Conservation Research.
“Now, we’re viewing life from an entirely new perspective. DNA carries instructions, and now we’re able to read those.”
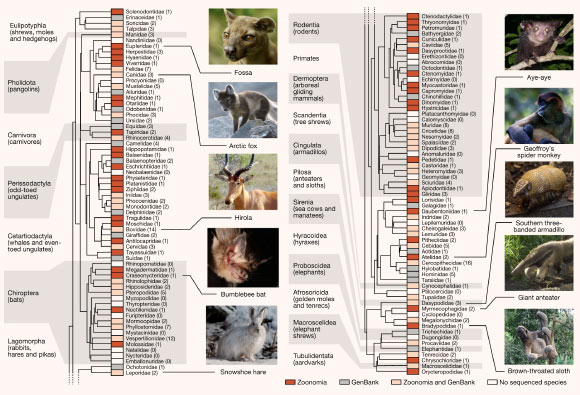
Phylogenetic tree of the mammalian families in the Zoonomia Project alignment, including both new assemblies and all other high-quality mammalian genomes publicly available in GenBank when the team started the alignment. Existing taxonomic classifications recognize a total of 127 extant families of eutherian mammal, including 43 families that were not previously represented in GenBank (red boxes) and 41 families with additional representative genome assemblies (pink boxes). Of the remaining families, 21 had GenBank genome assemblies but no Zoonomia Project assembly (gray boxes) and 22 had no representative genome assembly (white boxes). Parenthetical numbers indicate the number of species with genome assemblies in a given family. Image credit: Genereux et al., doi: 10.1038/s41586-020-2876-6.
In addition to understanding the human genome, all new placental mammal genomes together can be used to study how specific species adapt to different environments.
For example, some otters have a thick, water-resistant coat, and some mice, but not all, have adapted to hibernation. These animal traits can help us understand human traits such as metabolic diseases.
With climate change and more animal habitats being affected by human activities, it is becoming more and more important to defend endangered species.
In the new study, animals on the IUCN red list of endangered species had less variation in their genome, which is consistent with their endangered status.
“We hope that our extensive data set, which is available to all scientists in the world, will be used for understanding disease genetics and the protection of biodiversity,” Professor Lindblad-Toh said.
“Genome sequences for endangered species can help identify a species’ extinction risks and steer conservation efforts,” said Dr. Megan Owen, corporate director of wildlife conservation science at San Diego Zoo Global.
“They also give wildlife officials tools to apprehend poachers and wildlife traffickers.”
“One of the most exciting things about the Zoonomia Project is that many of our core questions are accessible to people both within and outside of science,” said Dr. Diane Genereux, a research scientist in the Vertebrate Genomics Group at the Broad Institute of MIT and Harvard.
“By designing scientific projects that are accessible to all, we can ensure benefits for public, human, and environmental health.”
The team’s results were published in the November 12, 2020 issue of the journal Nature.
_____
D.P. Genereux et al. (Zoonomia Consortium). 2020. A comparative genomics multitool for scientific discovery and conservation. Nature 587, 240-245; doi: 10.1038/s41586-020-2876-6







