The International Barley Genome Sequencing Consortium, which is led by Dr. Nils Stein of the Leibniz Institute of Plant Genetics and Crop Plant Research, Germany, has mapped the entire genome of barley (Hordeum vulgare), the world’s fourth most important cereal crop after wheat, rice and maize. The research appears in the journal Nature.
Barley was one of the first domesticated cereal grains, originating in the Fertile Crescent over 10,000 years ago.
It is widely cultivated in all temperate regions from the Arctic Circle to the tropics. In addition to its geographic adaptability, barley is particularly noted for its tolerance to cold, drought, alkali, and salinity.
Barley is found in breakfast cereals and all-purpose flour and helps bread rise. Malted barley gives beer color, body, protein to form a good head, and the natural sugars needed for fermentation.
The new research provides insights into gene families that are key to the malting process.
The barley genome sequence also enabled the identification of regions of the genome that have been vulnerable to genetic bottlenecking during domestication, knowledge that helps to guide breeders to optimize genetic diversity in their crop improvement efforts.
“It makes it much easier for researchers working with barley to be focused on attainable objectives, ranging from new variety development through breeding to mechanistic studies of genes,” said co-author Timothy J. Close, a professor of genetics at the University of California, Riverside.
“The research will also aid scientists working with other cereal crops.”
About 10 years ago, the International Barley Genome Sequencing Consortium set out to assemble a complete reference sequence of the barley genome.
This was a daunting task, as the barley genome is almost twice the size of the human genome and 80% of it is composed of highly repetitive sequences, which cannot be assigned accurately to specific positions in the genome without considerable extra effort.
Multiple novel strategies were used in this paper to circumvent this fundamental limitation. Major advances in sequencing technology, algorithmic design and computing made it possible.
“The barley genome is one of the largest in cereal crops and twice the size of the human genome,” the researchers explained.
“Barley is a true diploid, thus, it is a natural archetype for genetics and genomics for the Triticeae tribe, including polyploid wheat, and rye.”
This research provides knowledge of 39,734 high-confidence barley genes.
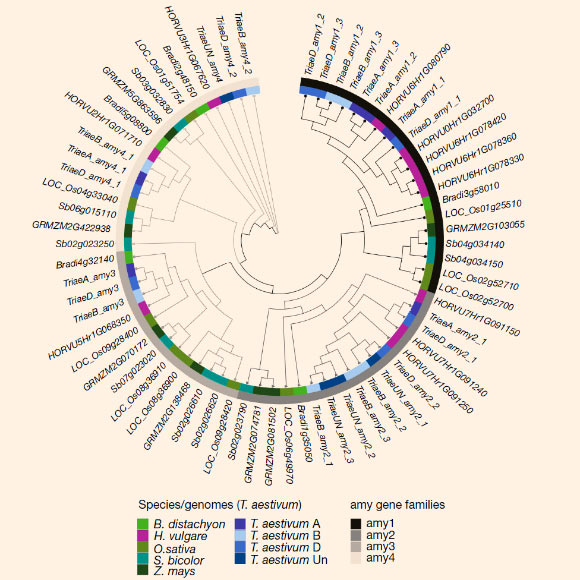
Phylogenetic tree of 68 full-length α-amylase protein sequences derived from amy genes identified in the genomes of barley, hexaploid wheat, B. distachyon, rice, sorghum and maize. Image credit: Mascher et al, doi: 10.1038/nature22043.
“Alcoholic beverages have been made from malted barley since the Stone Age, and some even consider this to be a major reason why humankind adopted plant cultivation, at least in the Fertile Crescent,” Prof. Close and co-authors said.
“During malting, amylase proteins are produced by germinated seeds to decompose energy-rich starch that is stored in dry grains, yielding simple sugars. These sugars then are available for fermentation by yeast to produce alcohol,” they explained.
“The genome sequence revealed much more variability than was expected in the genes that encode the amylase enzymes.”
_____
Martin Mascher et al. 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427-433; doi: 10.1038/nature22043