After decades of research aiming to understand how DNA is organized in our cells, a team of researchers from the Gladstone Institutes, the Massachusetts Institute of Technology and elsewhere has shed new light on this field by discovering how a key protein helps control gene organization. The study is published in the journal Cell (bioRxiv.org preprint).
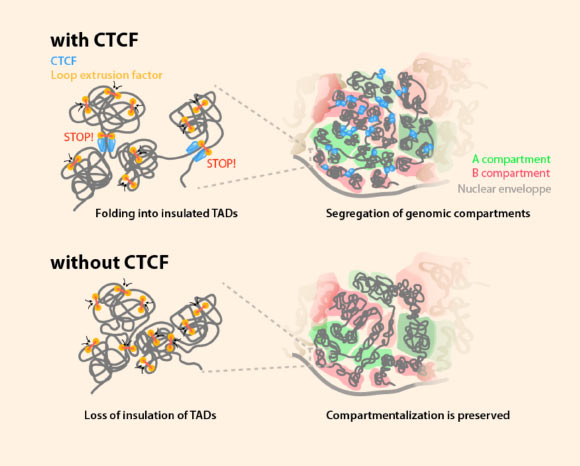
A study by Nora et al provides new insights into the rules governing human genome organization. Image credit: Nora et al, doi: 10.1016/j.cell.2017.05.004.
Humans have nearly 30,000 genes that determine traits from eye color to risk for hereditary diseases. Those genes sit along six feet of DNA, which are organized into chromosomes and stuffed into each and every human cell.
Chromosomes are coiled into loops and then organized into many large domains called topologically associating domains (TADs).
Within each TAD, several genes and the elements that regulate them are packaged together, and they are insulated from those in neighboring TADs.
“Imagine TADs are like adjoining rooms: like the genes in each TAD, people in each room can talk to one another, but not to people in the next room,” said study first author Dr. Elphège Nora, a postdoctoral researcher at the Gladstone Institute of Cardiovascular Disease.
“In previous work, we showed that TADs package genes together and insulate them from neighboring genes. The burning question then became: what controls this TAD organization?”
In the new study, Dr. Nora and co-authors discovered that the key to organizing these TADs is a protein called CTCF (also known as 11-zinc finger protein or CCCTC-binding factor).
“CTCF is a fascinating protein. It can be found at the boundaries of TAD domains, and was previously thought to be involved in many aspects of chromosome organization,” said senior author Dr. Benoit Bruneau, a professor at the University of California, San Francisco, and a senior investigator at the Gladstone Institute of Cardiovascular Disease.
“We wanted to see what would happen to the structure of chromosomes if we removed all the CTCF from cells.”
Scientists have struggled with studying the role of CTCF in the past, because it is absolutely essential to cells’ survival. Therefore, completely removing CTCF would cause cells to die, making them impossible to study.
“We used a new genetic method to completely eliminate CTCF in mammalian cells,” Dr. Nora explained.
“Using this technique, we destroyed the protein very quickly so that we could study the cells before they died. This allowed us to look at the entire genome in the absence of CTCF and observe the effects.”
The team demonstrated the importance of CTCF for the insulation of TADs.
“We noticed that, in the absence of the CTCF protein, the insulating boundaries of TAD domains had almost fully disappeared, so that genes and regulatory elements could now interact with those in adjacent TADs,” Dr. Nora said.
“This would be like removing the wall between adjoining rooms, so that people could now freely interact with others in the neighboring room.”
However, the absence of CTCF had little effect on how genes connect within a single TAD.
This indicates that CTCF is required for insulating TADs from one another, but not for packaging genes within these domains.
This represents the first conclusive study to show that the two mechanisms are separate and controlled by different proteins.
Now that the authors had a way of removing CTCF from cells, and disrupting the organization of TADs, they could start studying its impact on various aspects of the genome.
They leveraged this new ability to examine other levels of chromosome organization.
“We looked at a level of organization called compartmentalization, which separates active and inactive genes within a cell nucleus,” Dr. Nora said.
“This helps the cell identify which genes to use. For example, skins cells don’t need eye-related genes, so these genes would be tightly packaged in a compartment and put away, because the cell will never use them. We used to think that boundaries of TAD domains were a prerequisite for the organization of these compartments.”
“To our surprise, we found that is not the case. When we deleted the CTCF protein, which caused TAD boundaries to disappear, we saw no effect on the organization of the larger compartments,” Dr. Bruneau said.
“This interesting finding revealed that CTCF and TAD structure are not required for compartmentalization but, rather, that an independent mechanism is responsible for this chromosome organization.”
“Our findings redefine the role of CTCF in gene regulation and provide new insights about the fundamental processes that govern genome organization” Dr. Bruneau said.
“With this knowledge, we can now start reevaluating the cause of several diseases, as chromosome organization — including TADs — is often disrupted in many cancers and involved in significant developmental defects, such as congenital heart disease.”
_____
Elphège P. Nora et al. 2017. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169 (5): 930-944; doi: 10.1016/j.cell.2017.05.004
This article is based on text provided by the Gladstone Institutes.