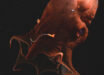
Scientists from the Okinawa Institute of Science and Technology Graduate University and elsewhere have produced a near-complete, haplotype-phased, genome assembly for the pearl oyster (Pinctada fucata).

The pearl oyster (Pinctada fucata). Image credit: K. Mikimoto & Co., Ltd / Pearl Research Institute.
“It’s very important to establish the genome,” said lead author Dr. Takeshi Takeuchi, a researcher in the Marine Genomics Unit at the Okinawa Institute of Science and Technology Graduate University.
“Genomes are the full set of an organism’s genes, many of which are essential for survival.”
“With the complete gene sequence, we can do many experiments and answer questions around immunity and how the pearls form.”
Pearl oysters receive special attention from researchers because of their value in the fishery industry for pearl production.
Pearls are one of the most beautiful jewels, largely produced in Japan since technical innovations in their cultivation in the early 20th century.
For the last decade, natural conditions at sites of pearl oyster fisheries have become less hospitable for reasons as yet not understood.
In the 1990s in Japan, mass mortality induced by infectious diseases reduced pearl production to about one third over the course of a decade.
Recently, fisheries have suffered disease outbreaks in juvenile and adult pearl oysters that may be related to both pathogens and increased seawater temperatures.
In 2012, Dr. Takeuchi and colleagues published a draft genome of the pearl oyster, which was one of the first molluscan draft genomes. Subsequently, they improved the draft assembly by producing more sequence data.
“These initial genome reconstructions were useful in resolving biological questions such as the population structure of the species,” they explained.
“However, to accurately understand the complexity of the pearl oyster genome and for aquaculture improvement, a more contiguous, high-quality genome resource with haplotype information is required.”
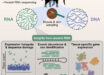
In their new research, Dr. Takeuchi and co-authors produced a fully phased assembly of the pearl oyster genome.
“Traditionally, when a genome is sequenced, researchers merge the pair of chromosomes together,” they said.
“This works well for lab animals, which normally have almost identical genetic information between the pair of chromosomes.”
“But for wild animals, where a considerable number of variants in genes exist between chromosome pairs, this method leads to a loss of information.”
“In our study, we decided not to merge the chromosomes when sequencing the genomes. Instead, we sequenced both sets of chromosomes — a method that is very uncommon. In fact, it’s probably the first research focused on marine invertebrates to use this method.”
“As pearl oysters have 14 pairs of chromosomes, they have 28 in total.”
The team reconstructed all 28 chromosomes and found key differences between the two chromosomes of one pair — chromosome pair 9. Notably, many of these genes were related to immunity.
“Different genes on a pair of chromosomes is a significant find because the proteins can recognize different types of infectious diseases,” Dr. Takeuchi said.
“When the animal is cultured, there is often a strain that has a higher rate of survival or produces more beautiful pearls.”
“The farmers often breed two animals with this strain but that leads to inbreeding and reduces genetic diversity.”
“We found that after three consecutive inbreeding cycles, the genetic diversity was significantly reduced.”
“If this reduced diversity occurs in the chromosome regions with genes related to immunity, it can impact the immunity of the animal.”
“It is important to maintain the genome diversity in aquaculture populations.”
The results appear in the journal DNA Research.
_____
Takeshi Takeuchi et al. 2022. A high-quality, haplotype-phased genome reconstruction reveals unexpected haplotype diversity in a pearl oyster. DNA Research 29 (6): dsac035; doi: 10.1093/dnares/dsac035