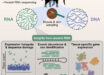
Scientists have sequenced the chromosome-scale genome of the apple cultivar ‘Honeycrisp,’ which has superior fruit quality traits, cold hardiness, and disease resistance.
Apples (Malus domestica) are the most consumed fruit in the United States.
The annual estimated total value of the U.S. apple industry is $23 billion, with five cultivars alone accounting for two-thirds of production (in order of proportion): ‘Gala,’ ‘Red Delicious,’ ‘Honeycrisp,’ ‘Granny Smith,’ and ‘Fuji.’
Of these, ‘Honeycrisp’ is by far the most valuable: it has roughly twice the value per pound of the next most valuable cultivar, ‘Fuji.’
‘Honeycrisp’ is appreciated by consumers, and therefore by the U.S. apple industry, for its superior flavor and crisp, juicy texture.
Importantly, properly stored ‘Honeycrisp’ fruit can be well-preserved for several months. Additionally, this cultivar shows high levels of cold hardiness and resistance to apple scab, the most economically important fungal disease of apples worldwide.
‘Honeycrisp’ was bred at the University of Minnesota in the 1960s, where the aim was to obtain cold hardy cultivars with high-quality fruit; it was released in 1991.
Recent genome-wide analysis (following the resolution of the ‘Honeycrisp’ pedigree) showed that the genetic background of ‘Honeycrisp’ is distinct from other important apple cultivars in the United States.
This is highlighted by the success of ‘Honeycrisp’ as a source of interesting genetic diversity in apple breeding programs worldwide to enhance texture, storability, and improved disease resistance. In fact, nine new cultivars derived from ‘Honeycrisp’ are already on the market.
To maximize both the understanding of genetic mechanisms driving important ‘Honeycrisp’ traits, and to assist tree fruit breeders, Cornell AgriTech researcher Awais Khan and colleagues aimed to sequence the high quality genome of this cultivar.
“Although it has many positive traits, it’s one of the most difficult apple cultivars to grow in the production system in orchards; it suffers from many physiological and post-harvest issues,” Dr. Khan said.
“For starters, Honeycrisp trees have difficulty getting enough nutrients on their own and require a specific nutrient management program for good yields and health.”
“Without such management, the trees commonly develop zonal leaf chlorosis, where leaves yellow and curl due to carbohydrate and nutrient imbalances.”
“Honeycrisp apples are also susceptible to disorders such as bitter pit, due to calcium imbalances, and bitter rot, a fungal infection.”
“Such issues are fundamentally genetically controlled, though improper handling and post-harvest storage can make them worse.”
“If we don’t know the genome and the genes in Honeycrisp, then we cannot specifically target and select for favorable traits and select out unfavorable traits through breeding.”
The apple genome is complex, large and heterozygous, meaning there are many versions of specific genes. There are also a lot of repeated sequences in the apple genome.
In 2010, when the first apple genome, of the Golden Delicious cultivar, was published, technologies could read only short fragments of DNA at a time, say, 150 letters.
Scientists would then overlap sequences of perhaps 50 letters, and like a puzzle, they would use computational programs and algorithms to match the end of one reading with the start of another. This allowed them to piece together longer strings of DNA to identify entire genes and eventually the genome.
But one problem with this method is that repeated elements can confuse the process.
In the new study, Dr. Khan and co-authors used a combination of current sequencing technologies — called PacBio HiFi, Omni-C and Illumina — that translated long reads of genetic sequences.
“We can sequence the whole larger fragment of the DNA sequence continuously, so we don’t have these big challenges of computational biology or bioinformatics to assemble and find the overlapping sequences,” Dr. Khan said.
“The long-read sequencing also helped us tease apart the apple’s diploid genome; like humans, apples have two sets of chromosomes, one from each parent.”
“The new technologies allowed us to sequence two single sets of chromosomes, which in future work may be used to differentiate between specific genetic contributions of each parent.”
The team’s work appears in the journal Gigabyte.
_____
Awais Khan et al. A phased, chromosome-scale genome of ‘Honeycrisp’ apple (Malus domestica). Gigabyte, published online September 19, 2022; doi: 10.46471/gigabyte.69