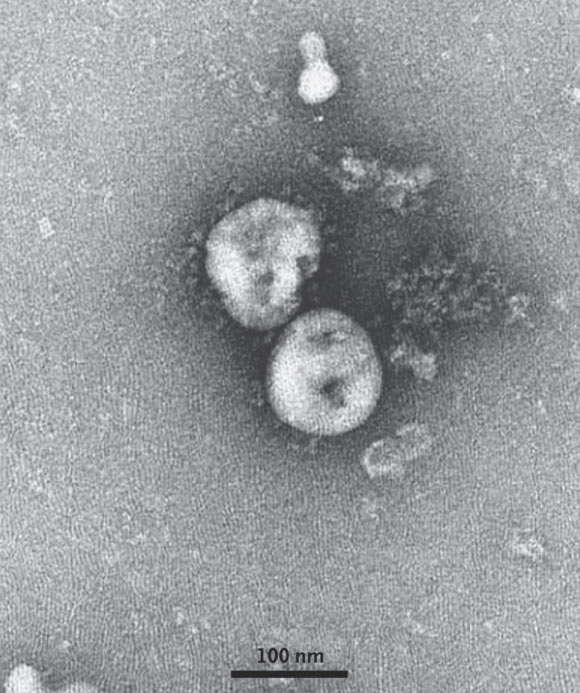
In a new study, a team of researchers from the University of Minnesota and the University of North Carolina has analyzed the potential receptor usage by the novel 2019-nCoV coronavirus, based on the rich knowledge about SARS-CoV (severe acute respiratory syndrome) and the recently released sequence of the new virus.
“The recent emergence of 2019-nCoV puts the world on alert,” said senior author Dr. Fang Li from the Department of Veterinary and Biomedical Sciences at the University of Minnesota and colleagues.
“2019-nCoV is reminiscent of the SARS-CoV outbreak in 2002-2003.”
“Our decade-long structural studies have shown how SARS-CoV interacts with animal and human hosts in order to infect them. The mechanics of infection by 2019-nCoV appear to be similar.”
Dr. Li and co-authors used the knowledge they gleaned from multiple strains of SARS-CoV — isolated from different hosts in different years — and angiotensin-converting enzyme-2 (ACE2) receptors from different animal species to model predictions for the new coronavirus.
Both viruses use ACE2 to gain entry into the cell, but it serves normally as a regulator for heart function.
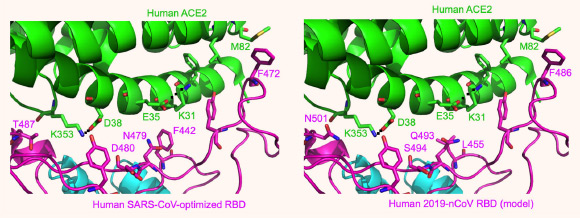
Left: experimentally determined structure of the interface between a designed SARS-CoV receptor-binding domain (RBD) — optimized for human ACE2 recognition — and human ACE2. Right: modeled structure of the interface between 2019-nCoV RBD and human ACE2; here mutations were introduced to the RBD region in left panel based on sequence differences between SARS-CoV and 2019-nCoV. Image credit: Wan et al, doi: 10.1128/JVI.00127-20.
“Our structural analyses confidently predict that 2019-nCoV uses ACE2 as its host receptor,” the researchers said.
“That and several other structural details of the new virus are consistent with the ability of the new coronavirus to infect humans and with some capability to transmit among humans.”
“Alarmingly, our data predict that a single mutation — at a specific spot in the genome — could significantly enhance 2019-nCoV’s ability to bind with human ACE2.”
“For this reason, 2019-nCoV evolution in patients should be closely monitored for the emergence of novel mutations at the 501 position in its genome, and to a lesser extent, the 494 position, in order to predict the possibility of a more serious outbreak than has been seen so far.”
The findings were published in the Journal of Virology, a journal of the American Society for Microbiology.
_____
Yushun Wan et al. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. Journal of Virology, published online January 29, 2020; doi: 10.1128/JVI.00127-20