An international team of researchers from Australia, China and the United States has shown for the first time that RNA interference is active in the response of human cells to some important viruses, including influenza A virus. The research is published in the journal Nature Microbiology.
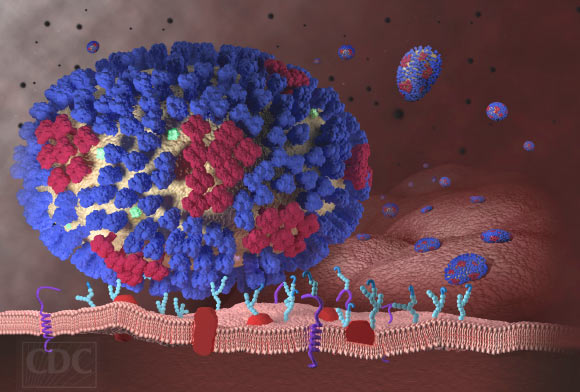
This image illustrates the beginning stages of an influenza infection and shows what happens after the influenza viruses enter the human body. Image credit: Centers for Disease Control and Prevention.
“Viruses are the most abundant infectious agents and are a constant threat to human health,” said co-corresponding author Dr. Kate Jeffrey, an investigator in the Massachusetts General Hospital gastrointestinal unit and an assistant professor of medicine at Harvard Medical School.
“Vaccines are somewhat effective but can have limited use when viruses like influenza rapidly mutate from year to year.”
“Identifying therapeutic targets within patients that could help them fight off an infection is a critical strategy for combating the spread of common, often-dangerous viruses.”
First described in the 1990s — a discovery that led to the 2006 Nobel Prize — RNA interference (RNAi) is a process by which organisms suppress the expression of target genes through the action of small RNA segments that bind to corresponding gene sequences.
Not only is RNAi used to regulate gene expression within an organism, it also can combat viral infection by silencing the activity of viral genes required for the pathogen’s replication.
Whether or not RNAi contributes to antiviral defense in humans and other mammals has been uncertain.
The only previous demonstration — by a research team led by University of California, Riverside Professor Shou-Wei Ding — was done in embryonic stem cells and in newborn mice.
Prof. Ding, who is co-corresponding author of the current study, has been studying antiviral RNAi for more than 20 years and also was the first to describe the action of the influenza virus protein NS1 (non-structural protein 1) in blocking RNAi in fruit flies, a common model system used by researchers.
In the current study, Prof. Ding, Dr. Jeffrey and their colleagues demonstrated that human cells produce abundant siRNAs (small interfering RNAs) to target the influenza A virus when the viral NS1 is not active.
The researchers showed that the creation of viral siRNAs in infected human cells is mediated by an enzyme known as Dicer and is potently suppressed by both the NS1 protein of influenza A virus and a protein (virion protein 35, or VP35) found in Ebola and Marburg viruses.
They further demonstrated that the infections of mature mammal cells by influenza A virus and other RNA viruses are inhibited naturally by RNAi, using mice cells specifically defective in RNAi.
“This opens up a new way to understand how humans respond to viral infections and develop new methods to control viral infections,” Prof. Ding said.
“Our studies show that the antiviral function of RNAi is conserved in mammals against distinct RNA viruses,” Dr. Jeffrey said.
“We now need to assess more directly the role of antiviral RNAi in human infectious diseases caused by RNA viruses — which include Ebola, West Nile and Zika along with influenza — and how harnessing or boosting the antiviral RNAi response could be used to reduce the severity of these infections,” she added.
_____
Yang Li et al. 2016. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nature Microbiology 2, article number: 16250; doi: 10.1038/nmicrobiol.2016.250







