A group of scientists at Boston College, Chestnut Hill, has reconstructed the natural history of a specific retrovirus lineage — ERV-Fc — that disseminated widely between 33 and 15 million years ago (Oligocene and early Miocene).
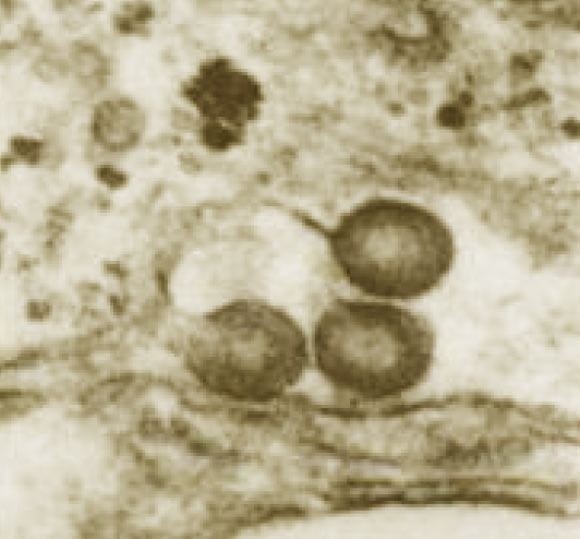
Retrovirus particles budding from rhesus macaque placenta cells. Image credit: Dorothy Feldman, via aacrjournals.org.
Retroviruses are abundant in nature and include human immunodeficiency viruses (HIV-1 and HIV-2), human T-cell leukemia viruses (HTLV-1 and -2), and the well-studied oncogenic retroviruses of mice and other model organisms, among many others.
The team’s findings, reported online on March 8, 2016, in the journal eLife, show that an ancient group of retroviruses known as ERV-Fc infected the ancestors of at least 28 mammal species — including carnivores, rodents and primates — between 15 million and 33 million years ago.
The distribution of ERV-Fc retroviruses among these ancient mammals suggests the viruses spread to every continent except Antarctica and Australia, and that they jumped from one species to another more than 20 times.
The study also places the origins of ERV-Fc at least as far back as the beginning of Oligocene.
“Unfortunately, viruses do not leave fossils behind, meaning we know very little about how they originate and evolve,” said team member Prof. Welkin Johnson, from the Boston College’s Biology Department.
“Over the course of millions of years, however, viral genetic sequences accumulate in the DNA genomes of living organisms, including humans, and can serve as molecular ‘fossils’ for exploring the natural history of viruses and their hosts.”
Using such ‘fossil’ remnants, Prof. Johnson and co-authors sought to uncover the natural history of ERV-Fc.
They were especially curious to know where and when these pathogens were found in the ancient world, which species they infected, and how they adapted to their mammalian hosts.
To do this, they first performed an exhaustive search of mammalian genome sequence databases for ERV-Fc loci and then compared the recovered sequences.
For each genome with sufficient ERV-Fc sequence, they reconstructed the sequences of proteins representing the virus that colonized the ancestors of that particular species.
These sequences were then used to infer the natural history and evolutionary relationships of ERV-Fc-related viruses.
“Mammalian genomes contain hundreds of thousands of ancient viral fossils similar to ERV-Fc,” the scientists said.
“Future work could study these to improve our understanding of when and why new viruses emerge and how long-term contact with viruses affects the evolution of their host organisms.”
_____
William E. Diehl et al. 2016. Tracking interspecies transmission and long-term evolution of an ancient retrovirus using the genomes of modern mammals. eLife 5: e12704; doi: 10.7554/eLife.12704