A team of researchers from the Okinawa Institute of Science and Technology Graduate University and University College London has decoded the genome of Bruguiera gymnorhiza, one of the most widely distributed mangrove trees from the most mangrove-rich family Rhizophoraceae. The new data provide novel insights into the epigenome regulation of mangroves and a better understanding of the adaptation of plants to fluctuating, harsh natural environments.
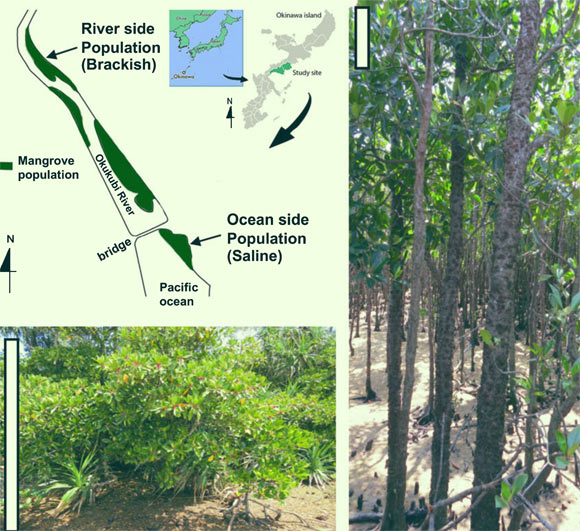
Representative Bruguiera gymnorhiza trees in the river side population (left) and the oceanside population (right) along the Okukubi River, the Okinawa main island, Japan. Scale bars – 1 m. Image credit: Miryeganeh et al., doi: 10.1111/nph.17738.
Mangroves are an important ecosystem for the planet, protecting coastlines from erosion, filtering out pollutants from water and serving as a nursery for fish and other species that support coastal livelihoods.
They also play a crucial role in combating global warming, storing up to four times as much carbon in a given area as a rainforest.
Despite their importance, mangroves are being deforested at an unprecedented rate, and due to human pressure and rising seas, are forecast to disappear in as little as 100 years.
“Mangroves are an ideal model system for studying the molecular mechanism behind stress tolerance, as they naturally cope with various stress factors,” said study first author Dr. Matin Miryeganeh, a researcher in the Plant Epigenetics Unit at the Okinawa Institute of Science and Technology Graduate University.
During a survey of mangrove trees in Okinawa, Japan, the authors noticed that Bruguiera gymnorhiza trees showed striking differences between individuals rooted in the oceanside, with high salinity, and those in the upper riverside, where the waters were more brackish.
“The trees were amazingly different: near the ocean, the height of the trees was about 1-2 m, whereas further up the river, the trees grew as high as 7 m,” said study senior author Professor Hidetoshi Saze, head of the Plant Epigenetics Unit at the Okinawa Institute of Science and Technology Graduate University.
“But the shorter trees were not unhealthy — they flowered and fruited normally — so we think this modification is adaptive, perhaps allowing the salt-stressed plant to invest more resources into coping with its harsh environment.”
In their research, the scientists sequenced the genome of Bruguiera gymnorhiza and found that it contained 309 million base pairs.
They then identified a total of 34,403 genes — a much larger genome than those for other known mangrove tree species.
The large size was due to, for the most part, almost half of the DNA being made up of repeating sequences.
When the researchers examined the type of repetitive DNA, they found that over a quarter of the genome consisted of genetic elements called transposons, or ‘jumping genes.’
“Active transposons are parasitic genes that can ‘jump’ position within the genome, like cut-and paste or copy-and-paste computer functions,” Professor Saze said.
“As more copies of themselves are inserted into the genome, repetitive DNA can build up.”
The team then examined how activity of Bruguiera gymnorhiza’s genes, including the transposons, varied between individuals in the oceanside location with high salinity, and individuals in the less saline, brackish waters upriver.
They also compared gene activity for mangrove trees grown in the lab, under two different conditions that replicated the oceanside and upriver salinity levels.
Overall, in both the oceanside individuals and those grown in high salinity conditions in the lab, genes involved in suppressing transposon activity showed higher expression, while genes that normally promote transposon activity showed lower expression.
In addition, when the authors looked specifically into transposons, they found evidence of chemical modifications on their DNA that lowered their activity.
“This shows that an important means of coping with saline stress involves silencing transposons,” Dr. Miryeganeh said.
The scientists also saw increases in the activity of genes involved in stress responses in plants, including those that activate when plants are water-deprived.
Gene activity also suggested the stressed plants have lower levels of photosynthesis.
“This study acts as a foundation, providing new insights into how mangrove trees regulate their genome in response to extreme stresses,” Professor Saze said.
“More research is needed to understand how these changes in gene activity impact molecular processes within the plant cells and tissues and could one day help scientists create new plant strains that can better cope with stress.”
A paper on the findings was published in the journal New Phytologist.
_____
Matin Miryeganeh et al. De novo genome assembly and in natura epigenomics reveal salinity-induced DNA methylation in the mangrove tree Bruguiera gymnorhiza. New Phytologist, published online September 16, 2021; doi: 10.1111/nph.17738