Scientists from the Max Planck Institute for Plant Breeding Research and elsewhere have decoded the genome of the potato cultivar ‘Otava.’ Their results shed new light on the recent breeding history of potato, the functional organization of its tetraploid genome and has the potential to strengthen the future of genomics-assisted breeding.
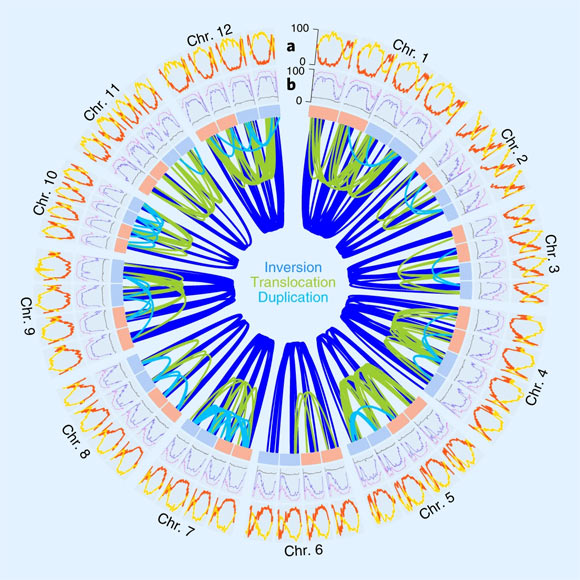
The genomic features of the genome of the autotetraploid potato (Solanum tuberosum): (a) gene density (number of genes per Mb; red) and percentage of TE-related sequence (yellow) within 2-Mb windows along the four haplotypes of each of the 12 chromosomes (Chr. 1-12); (b) landscapes of methylation in CG (purple), CHG (blue), CHH (gray) context within 2-Mb windows; the links in the center show >600 structural rearrangements >100 kb found between the four haplotypes of each chromosome; light blue and red boxes refer to the maternally and paternally inherited chromosomes. Image credit: Sun et al., doi: 10.1038/s41588-022-01015-0.
Potato (Solanum tuberosum) is a member of the Solanaceae, an economically important family that includes tomato, pepper, aubergine (eggplant), petunia and tobacco.
It is an important tuber crop and is among the five most produced crops in the world.
Globally more than 350 million tons of potato are produced per year with an increasing trend particularly in developing countries in Asia.
Despite the social and economic importance, the breeding success of potato remained low over the past decades due to its heterozygous, autotetraploid genome and the high levels of inbreeding depression, which challenge usual breeding strategies commonly applied to inbred, diploid crops.
A fundamental tool for modern breeding is the availability of reference sequences.
The reference sequence for potato was generated from a double haploid plant, was initially published in 2011, and continuously improved over the past years, including a recent update based on long read sequencing.
Another major advancement in potato genomics was the recent assembly of a heterozygous diploid potato. This haplotype-resolved genome was generated from a variety of different sequencing technologies and phase information from a genetic map derived from selfed progeny.
However, as of now, there is no haplotype-resolved assembly of a tetraploid potato cultivar available nor is there a straightforward method that would enable the assembly of the individual haplotypes of a tetraploid genome.
“The potato is becoming more and more integral to diets worldwide including even Asian countries like China where rice is the traditional staple food,” said study senior author Professor Korbinian Schneeberger, a researcher in the Faculty of Biology at the LMU Munich and the Department of Chromosome Biology at the Max Planck Institute for Plant Breeding Research.
“Building on this new work, we can now implement genome-assisted breeding of new potato varieties that will be more productive and also resistant to climate change — this could have a huge impact on delivering food security in the decades to come.”
In the research, Professor Schneeberger and colleagues generated an assembly of the autotetraploid genome of the potato cultivar ‘Otava.’
“We report the 3.1 Gb haplotype-resolved (at 99.6% precision), chromosome-scale assembly of the potato cultivar ‘Otava’ based on high-quality long reads, single-cell sequencing of 717 pollen genomes and Hi-C data,” they said.
“Unexpectedly, roughly 50% of the genome was identical-by-descent due to recent inbreeding, which was contrasted by highly abundant structural rearrangements involving about 20% of the genome.”
“Among 38,214 genes, only 54% were present in all four haplotypes with an average of 3.2 copies per gene.”
“Taking the leaf transcriptome as an example, 11% of the genes were differently expressed in at least one haplotype, where 25% of them were likely regulated through allele-specific DNA methylation.”
“Our work sheds light on the recent breeding history of potato, the functional organization of its tetraploid genome and has the potential to strengthen the future of genomics-assisted breeding.”
The results were published March 3, 2022 in the journal Nature Genetics.
_____
H. Sun et al. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nat Genet, published online March 3, 2022; doi: 10.1038/s41588-022-01015-0