Using the NSF-funded Frontera supercomputer at the University of Texas at Austin’s Texas Advanced Computing Center (TACC), researchers are preparing a massive computer model of SARS-CoV-2, a novel coronavirus which causes the COVID-19 disease, that they expect will give insight into how it infects in the human body.
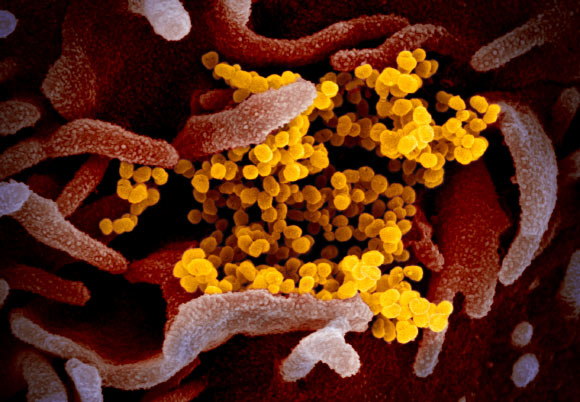
This scanning electron microscope image shows COVID-19 virus (yellow), also known as 2019-nCoV and SARS-CoV-2, isolated from a patient in the U.S., emerging from the surface of cells (pink) cultured in the lab. Image credit: NIAID-RML / CC BY 2.0.
“If we have a good model for what the outside of the particle looks like and how it behaves, we’re going to get a good view of the different components that are involved in molecular recognition,” said Professor Rommie Amaro, from the University of California, San Diego.
“Molecular recognition involves how the SARS-CoV-2 virus interacts with the angiotensin converting enzyme 2 (ACE2) receptors and possibly other targets within the host cell membrane.”
The coronavirus model is anticipated by the team to contain roughly 200 million atoms, a daunting undertaking, as the interaction of each atom with one another has to be computed.
“We’re trying to combine data at different resolutions into one cohesive model that can be simulated on leadership-class facilities like Frontera,” Professor Amaro said.
“We basically start with the individual components, where their structures have been resolved at atomic or near atomic resolution. We carefully get each of these components up and running and into a state where they are stable. Then we can introduce them into the bigger envelope simulations with neighboring molecules.”
The team’s work with the coronavirus builds on the success with an all-atom simulation of the influenza virus envelope, published in the journal ACS Central Science.
“The influenza work will have a remarkable number of similarities to what they’re now pursuing with the coronavirus,” Professor Amaro said.
“It’s a brilliant test of our methods and our abilities to adapt to new data and to get this up and running right off the fly.”
“It took us a year or more to build the influenza viral envelope and get it up and running on the national supercomputers. For influenza, we used the Blue Waters supercomputer, which was in some ways the predecessor to Frontera. The work, however, with the coronavirus obviously is proceeding at a much, much faster pace. This is enabled, in part because of the work that we did on Blue Waters earlier.”
“These simulations will give us new insights into the different parts of the coronavirus that are required for infectivity,” Professor Amaro added.
“And why we care about that is because if we can understand these different features, scientists have a better chance to design new drugs; to understand how current drugs work and potential drug combinations work.”
“The information that we get from these simulations is multifaceted and multidimensional and will be of use for scientists on the front lines immediately and also in the longer term.”
“Hopefully the public will understand that there’s many different components and facets of science to push forward to understand this virus.”
“These simulations on Frontera are just one of those components, but hopefully an important and a gainful one,” she said.