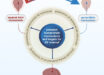
A revolutionary new technique combines a rapid 3D microscopy technique known as lattice light-sheet microscopy with expansion microscopy for nanoscale imaging of fruit fly (Drosophila melanogaster) and mouse neuronal circuits and their molecular constituents that’s roughly 1,000 times faster than other methods.

ExLLSM (expansion/lattice light-sheet microscopy) images neural structures with molecular contrast over millimeter-scale volumes, including (clockwise from top right) mouse pyramidal neurons and their processes; organelle morphologies in somata; dendritic spines and synaptic proteins across the cortex; stereotypy of projection neuron boutons in Drosophila; projection neurons traced to the central complex; and (center) dopaminergic neurons across the brain, including the ellipsoid body (circular inset). Image credit: Gao et al, doi: 10.1126/science.aau8302.
Lattice light-sheet microscopy uses highly focused light beams to rapidly assemble a 3D image of a specimen one thin slice at a time.
Expansion microscopy involves fixing tissue and then expanding it like a balloon while keeping the relative positions of internal structures unchanged. It uses a polyacrylamide gel like that in diapers, which swells when moved from salty to pure water.
“A lot of problems in biology are multiscale,” said MIT’s Professor Edward Boyden, a member of MIT’s McGovern Institute for Brain Research, Media Lab, and Koch Institute for Integrative Cancer Research.
“Using lattice light-sheet microscopy, along with the expansion microscopy process, we can now image at large scale without losing sight of the nanoscale configuration of biomolecules.”
“The marrying of the lattice light-sheet microscope with expansion microscopy is essential to achieve the sensitivity, resolution, and scalability of the imaging that we’re doing,” added Dr. Ruixuan Gao, a postdoctoral researcher at MIT.
Imaging expanded tissue samples generates huge amounts of data — up to tens of terabytes per sample — so the researchers also had to devise highly parallelized computational image-processing techniques that could break down the data into smaller chunks, analyze it, and stitch it back together into a coherent whole.
In the new study, they demonstrated the power of their new technique by imaging layers of neurons in the somatosensory cortex of mice, after expanding the tissue volume fourfold.
They focused on a type of neuron known as pyramidal cells, one of the most common excitatory neurons found in the nervous system.
To locate synapses, or connections, between these neurons, they labeled proteins found in the presynaptic and postsynaptic regions of the cells. This also allowed them to compare the density of synapses in different parts of the cortex.
Using this technique, it is possible to analyze millions of synapses in just a few days.
“We counted clusters of postsynaptic markers across the cortex, and we saw differences in synaptic density in different layers of the cortex. Using electron microscopy, this would have taken years to complete,” Dr. Gao said.
The researchers also studied patterns of axon myelination (myelin is a fatty substance that insulates axons and whose disruption is a hallmark of multiple sclerosis) in different neurons.
They were able to compute the thickness of the myelin coating in different segments of axons, and they measured the gaps between stretches of myelin, which are important because they help conduct electrical signals. Previously, this kind of myelin tracing would have required months to years for human annotators to perform.
This technology can also be used to image tiny organelles inside neurons. In the study, the scientists were able to identify mitochondria and lysosomes and also measure variations in the shapes of these organelles.
The researchers also demonstrated that this technique could be used to analyze brain tissue from other organisms as well.
They used it to image the entire brain of the fruit fly, which is the size of a poppy seed and contains about 100,000 neurons.
In one set of experiments, they traced an olfactory circuit that extends across several brain regions, imaged all dopaminergic neurons, and counted all synapses across the brain.
By comparing multiple animals, they also found differences in the numbers and arrangements of synaptic boutons within each animal’s olfactory circuit.
“In future work, this technique could be used to trace circuits that control memory formation and recall, to study how sensory input leads to a specific behavior, or to analyze how emotions are coupled to decision-making,” Professor Boyden said.
“These are all questions at a scale that you can’t answer with classical technologies.”
“The system could also have applications beyond neuroscience. We are planning to work with other researchers to study how HIV evades the immune system, and the technology could also be adapted to study how cancer cells interact with surrounding cells, including immune cells.”
The team’s work is published in the journal Science.
_____
Ruixuan Gao et al. 2019. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363 (6424); doi: 10.1126/science.aau8302