A team of researchers at Pennsylvania State University has directly observed functional metabolons — long-hypothesized clusters of enzymes — involved in generating purines (building blocks of DNA and RNA), the most abundant cellular metabolites.
“Our study suggests that enzymes are not haphazardly located throughout cells, but instead occur in discrete clusters, or metabolons, that carry out specific metabolic pathways,” said senior co-author Professor Stephen Benkovic, a researcher in the Department of Chemistry at Pennsylvania State University.
“Not only did we find proof that metabolons exist, but we also found that this metabolon occurs near mitochondria in cancer cells.”
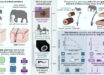
In the study, Professor Benkovic and colleagues searched for a purinosome, a specific kind of metabolon that was thought to carry out de novo purine biosynthesis (DNPB), the process by which new purines are synthesized.
The team investigated these purinosomes within HeLa cells, a cervical cancer cell line commonly used in scientific research.
“We have shown that the DNPB pathway is carried out by purinosomes consisting of at least nine enzymes acting together synergistically to increase their overall activity by at least by seven-fold,” said lead author Dr. Vidhi Pareek, a researcher in the Huck Institutes of Life Sciences at Pennsylvania State University.
The scientists identified the purinosomes, which were less than a micrometer in diameter, using a novel imaging system.
“The technique utilizes gas cluster ion beam secondary ion mass spectrometry (GCIB-SIMS) to detect intact biomolecules with high sensitivity and allow in situ chemical imaging in single cells,” said co-author Dr. Hua Tian, a researcher in the Department of Chemistry and the Materials Research Institute at Pennsylvania State University.
“This was vital for the study since we are dealing with very low concentration of molecules in individual cancer cells.”
“Now, at the end of my career, I am finally seeing this imaging approach reveal the presence of purinosomes, and perhaps next, observe that a cancer drug actually makes it into a purinosome where it can be most effective,” added senior co-author Professor Nicholas Winograd, from the Department of Chemistry at Pennsylvania State University.
Importantly, the study authors found that the DNPB pathway occurs in a channeled manner and the juxtaposition of purinosomes to the mitochondria facilitates uptake of substrates generated by the mitochondria for utilization in the pathway.
Channeling occurs when enzymes are located close together so that the molecules produced are quickly transferred and processed along the enzymatic pathway, restricting equilibration with the bulk cytosol.
“Our experiment allowed us to show that the efficiency of the DNPB pathway is increased by channeling and that the proximity of purinosomes near mitochondria is consequential for the pathway,” Professor Benkovic said.
“These findings open the door to the study of a new class of cancer therapeutics; for example the design of a molecule that can disrupt purinosomes’ juxtaposition with mitochondria.”
The study was published in the journal Science.
_____
Vidhi Pareek et al. 2020. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science 368 (6488): 283-290; doi: 10.1126/science.aaz6465