For the first time, researchers have used the CRISPR-Cas microbial immune system to encode a primitive digital movie into — and then ‘play it back’ from — the genomes of living E.coli bacteria.
DNA is emerging as an excellent medium for storing data. Image credit: Shipman et al, doi: 10.1038/nature23017.
“We want to turn cells into historians. We envision a biological memory system that’s much smaller and more versatile than today’s technologies, which will track many events non-intrusively over time,” said Dr. Seth Shipman, a postdoctoral fellow at Harvard Medical School and firs author of the paper reporting the results in the journal Nature.
The ability to record such sequential events like a movie at the molecular level is key to the idea of reinventing the very concept of recording using molecular engineering.
In this scheme, cells themselves could be induced to record molecular events – such as changes in gene expression over time — in their own genomes.
Then the information could be retrieved simply by sequencing the genomes of the cells it is stored in.
“If we had those transcriptional steps, we could potentially use them like a recipe to engineer similar cells. These could be used to model disease — or even in therapies,” Dr. Shipman said.
For starters, Dr. Shipman and co-authors had to show that DNA can be used to encode not just genetic information, but any arbitrary sequential information into a genome. For this they turned to the CRISPR-Cas9 gene editing tool.
They first demonstrated that they could encode and retrieve an image of the human hand in DNA inserted into E.coli bacteria.
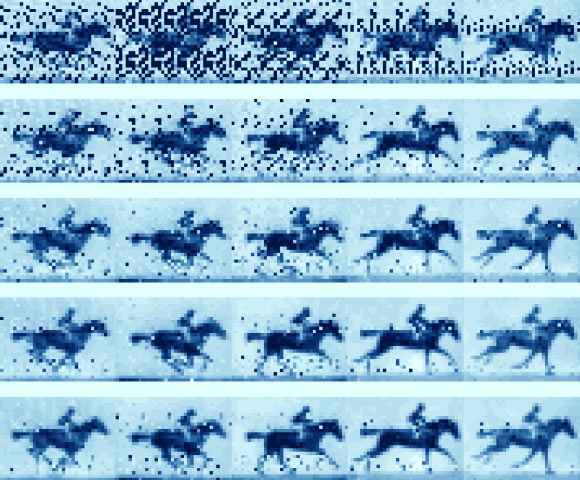
They then similarly encoded and reconstructed frames from a classic 1870s race horse in motion sequence of photos — an early forerunner of moving pictures.
“The sequential nature of CRISPR makes it an appealing system for recording events over time,” Dr. Shipman said.
The team then similarly translated five frames from the race horse in motion photo sequence into DNA.
Over the course of five days, the authors sequentially treated bacteria with a frame of translated DNA.
Afterwards, they were able to reconstruct the movie with 90% accuracy by sequencing the bacterial DNA.
“Although this technology could be used in a variety of ways, we ultimately hope to use it to study the brain,” the scientists said.
“We want to use neurons to record a molecular history of the brain through development,” Dr. Shipman said.
“Such a ‘molecular recorder’ will allow us to eventually collect data from every cell in the brain at once, without the need to gain access, to observe the cells directly, or disrupt the system to extract genetic material or proteins.”
_____
Seth L. Shipman et al. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature, published online July 12, 2017; doi: 10.1038/nature23017