Using a novel approach called syn-BNP (synthetic-bioinformatic natural product) approach, scientists from the Rockefeller and Rutgers Universities have discovered two promising new antibiotics active against methicillin-resistant Staphylococcus aureus (MRSA), a bacterium that causes infections in different parts of the body and poses a serious threat to millions of people throughout the world.
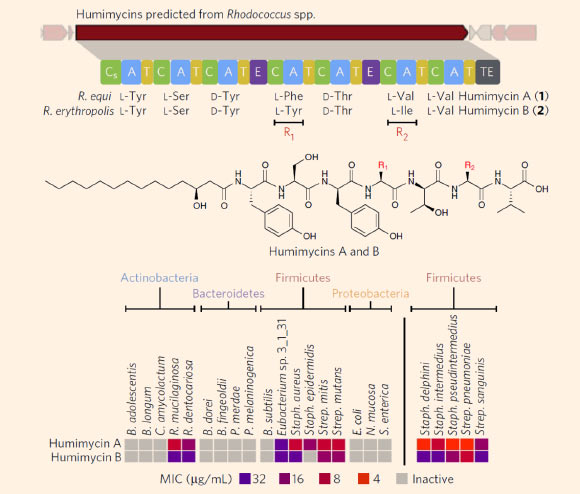
Discovery and screening of the humimycins. Top: the humimycins were predicted from closely related gene clusters found in two Rhodococcus spp. cultured from human subjects. Center: chemical structures of humimycin A and B; the two antibiotics differ only at the fourth (F or Y) and sixth (V or I) residues. Bottom: MIC values for the humimycins against a panel of human commensal and pathogenic bacteria; the humimycins were particularly active against bacteria in the Staphylococcus and Streptococcus genus. Image credit: John Chu et al.
“In our approach, natural product structures are bioinformatically predicted from primary sequence data and produced by chemical synthesis,” the authors explained.
Led by Rockefeller University researcher Sean Brady, the team began by trawling publicly available databases for the genomes of bacteria that reside in the human body.
They then used specialized computer software to scan hundreds of those genomes for clusters of genes that were likely to produce molecules known as non-ribosomal peptides that form the basis of many antibiotics.
They also used the software to predict the chemical structures of the molecules that the gene clusters ought to produce.
The software initially identified 57 potentially useful gene clusters, which Dr. Brady and co-authors winnowed down to 30. They then used a method called solid-phase peptide synthesis to manufacture 25 different chemical compounds.
By testing those compounds against human pathogens, the team successfully identified two closely related antibiotics, which they named humimycin A and humimycin B.
Both are found in Rhodococcus equi and R. erythropolis — microbes that had never yielded anything resembling the humimycins when cultured using traditional laboratory techniques.
The humimycins proved especially effective against Staphylococcus and Streptococcus bacteria, which can cause dangerous infections in humans and tend to grow resistant to various antibiotics.
Further experiments suggested that the humimycins work by inhibiting an enzyme that bacteria use to build their cell walls — and once that cell-wall building pathway is interrupted, the bacteria die.
A similar mode of action is employed by beta-lactams, a broad class of commonly prescribed antibiotics whose effect often wanes as bacteria develop ways to resist them.
Yet the team found that one of the humimycins could be used to re-sensitize bacteria to beta-lactams that they had previously outsmarted.
In one experiment, Dr. Brady and his colleagues exposed beta-lactam resistant Staphylococcus microbes to humimycin A in combination with a beta-lactam antibiotic, and the bugs once again succumbed.
Remarkably, that held true even when humimycin A had little effect by itself — a result that the authors attribute to the fact that both compounds work by interrupting different steps in the same biological pathway.
To further test that proposition, the team infected mice with a beta-lactam resistant strain of Staphylococcus aureus.
Mice that were subsequently treated with a mixture containing both humimycin A and a beta-lactam antibiotic fared far better than those treated with only one drug or the other — a finding that could point towards a new treatment regimen for humans infected with beta-lactam resistant S. aureus.
“We hope that this discovery will inspire scientists to mine the genomes of bacteria for more molecules that could yield similarly useful results,” the researchers said.
Details of the research were recently published in the journal Nature Chemical Biology.
_____
John Chu et al. 2016. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nature Chemical Biology 12: 1004-1006; doi: 10.1038/nchembio.2207