A multinational team of researchers led by University College London (UCL) has found four genes that impact nose shape.

Image showing variation between nose shape and the specific genes responsible. Image credit: Kaustubh Adhikari / University College London.
The four genes mainly affect the width and ‘pointiness’ of noses which vary greatly between different populations, according to the team, led by Prof. Andrés Ruiz-Linares of UCL Genetics Institute.
The scientists analyzed a population of over 6,000 people with varied ancestry across Latin America to study the differences in normal facial features and identify the genes which control the shape of the nose and chin.
Both men and women were assessed for 14 different facial features and whole genome analysis identified the genes driving differences in appearance.
“Our study sample is part of the CANDELA cohort collected in Latin America (Brazil, Colombia, Chile, Mexico and Peru),” they said. “Using facial photographs of 6,275 individuals, we assessed 14 facial features on an ordered categorical scale reflecting the distinctiveness of each trait.”
“We included features of the lower face: chin shape, chin protrusion and upper/lower lip thickness; the middle face: cheekbone protrusion, breadth of nasal root, bridge and wing, columella inclination, nose protrusion, nose profile and nose tip shape; and the upper face: brow-ridge protrusion and forehead profile. These features were selected based on their documented variation in Europeans.”
Prof. Ruiz-Linares and co-authors identified five genes which play a role in controlling the shape of specific facial features.
DCHS2, RUNX2, GLI3 and PAX1 affect the width and pointiness of the nose and another gene — EDAR — affects chin protrusion.
A subgroup of 3,000 individuals had their features assessed using a 3D reconstruction of the face in order to obtain exact measurements of facial features and the results identified the same genes.
According to the scientists, GLI3, DCHS2 and PAX1 are all genes known to drive cartilage growth — GLI3 gave the strongest signal for controlling the breadth of nostrils, DCHS2 was found to control nose pointiness and PAX1 also influences nostril breadth. RUNX2 which drives bone growth was seen to control nose bridge width.
The genes GLI3, DCHS2 and RUNX2 are known to show strong signals of recent selection in modern humans compared to archaic humans such as Neanderthals and Denisovans; GLI3 in particular undergoing rapid evolution.
“It has long been speculated that the shape of the nose reflects the environment in which humans evolved,” Prof. Ruiz-Linares said.
“For example, the comparatively narrower nose of Europeans has been proposed to represent an adaptation to a cold, dry climate. Identifying genes affecting nose shape provides us with new tools to examine this question, as well as the evolution of the face in other species. It may also help us understand what goes wrong in genetic disorders involving facial abnormalities.”
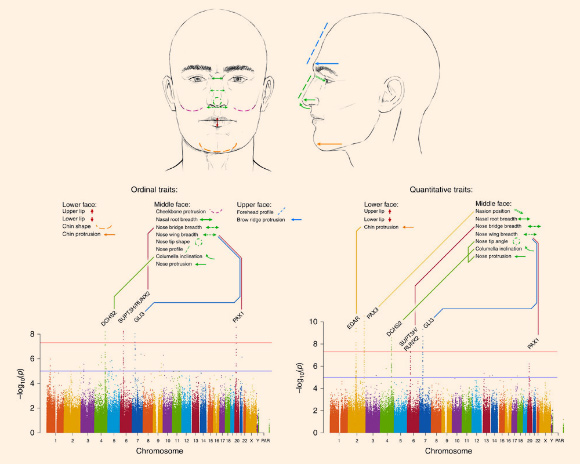
The team first carried out a genome-wide association study using data for 14 ordinal facial features from the lower, middle and upper face in 5,958 individuals. For follow-up, they obtained quantitative proxies for 9 of the 14 ordinal traits initially examined in a subset of 2,955 individuals, and performed another genome-wide association study. Image credit: Emiliano Bellini / Kaustubh Adhikari et al.
“Few studies have looked at how normal facial features develop and those that have only looked at European populations, which show less diversity than the group we studied,” added first author Dr. Kaustubh Adhikari, UCL Cell & Developmental Biology.
“What we’ve found are specific genes which influence the shape and size of individual features, which hasn’t been seen before.”
“Finding out the role each gene plays helps us to piece together the evolutionary path from Neanderthal to modern humans.”
“It brings us closer to understanding how genes influence the way we look, which is important for forensics applications.”
The results were published online this week in the journal Nature Communications.
_____
Kaustubh Adhikari et al. 2016. A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nature Communications 7, article number: 11616; doi: 10.1038/ncomms11616