U.S. researchers have discovered two large-effect mutations that sparked a hormonal revolution about 500 million years ago.
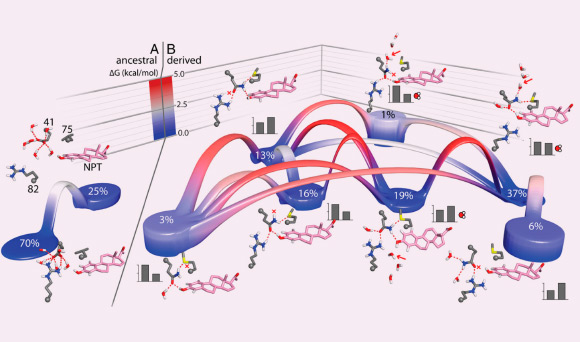
Two mutations altered the energetic landscape of protein-ligand binding. A, left: thermodynamic landscapes of the ensemble of interaction networks of the synthetic estrogen NPT and residues 41, 75, and 82 are shown for AncSR2 – the ancestor of the nonaromatized steroid receptors clade – containing the ancestral states glu41 and leu75; or, B, the derived states GLN41 and MET75 (adapted from Michael J. Harms et al., 2013)
In their study, reported in the Proceedings of the National Academy of Sciences, the scientists studied the evolution of a family of proteins called steroid hormone receptors, which mediate the effects of hormones on reproduction, development and physiology.
They traced how the ancestor of the entire receptor family – which recognized only estrogens – evolved into descendant proteins capable of recognizing other steroid hormones, such as testosterone, progesterone and the stress hormone cortisol.
To do so, the researchers used a gene ‘resurrection’ strategy. They first inferred the genetic sequences of ancient receptor proteins, using computational methods to work their way back up the tree of life from a database of hundreds of present-day receptor sequences.
They then biochemically synthesized these ancient DNA sequences and used molecular assays to determine the receptors’ sensitivity to various hormones.
The scientists narrowed down the time range during which the capacity to recognize non-estrogen steroids evolved, to a period about 500 million years ago, before the dawn of vertebrate animals on Earth.
They then identified the most important mutations that occurred during that interval by introducing them into the reconstructed ancestral proteins. By measuring how the mutations affected the receptor’s structure and function, they could re-create ancient molecular evolution in the laboratory.
They found that just two changes in the ancient receptor’s gene sequence caused a 70,000-fold shift in preference away from estrogens toward other steroid hormones.
“Changes in just two letters of the genetic code in our deep evolutionary past caused a massive shift in the function of one protein and set in motion the evolution of our present-day hormonal and reproductive systems,” said senior author Prof Joe Thornton from the University of Chicago.
Prof Thornton and his colleagues also used biophysical techniques to identify the precise atomic-level mechanisms by which the mutations affected the protein’s functions. Although only a few atoms in the protein were changed, this radically rewired the network of interactions between the receptor and the hormone, leading to a massive change in function.
“If those two mutations had not happened, our bodies today would have to use different mechanisms to regulate pregnancy, libido, the response to stress, kidney function, inflammation, and the development of male and female characteristics at puberty.”
“Our findings show that new molecular functions can evolve by sudden large leaps due to a few tiny changes in the genetic code. Along with the two key changes in the receptor, additional mutations, the precise effects of which are not yet known, were necessary for the full effects of hormone signaling on the body to evolve,” Prof Thornton said.
______
Bibliographic information: Michael J. Harms et al. Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. PNAS, published online before print June 24, 2013; doi: 10.1073/pnas.1303930110