Chromatin is a substance within a chromosome consisting of DNA and protein. The major proteins in chromatin are histones, which help package the DNA in a compact form that fits in the cell nucleus. New research from the Weizmann Institute of Science has revealed a previously unknown mode of nuclear chromatin organization in fully differentiated cells in which chromatin density is high at the nuclear periphery and undetectable in the nuclear center, creating an effectively central chromatin-devoid region.
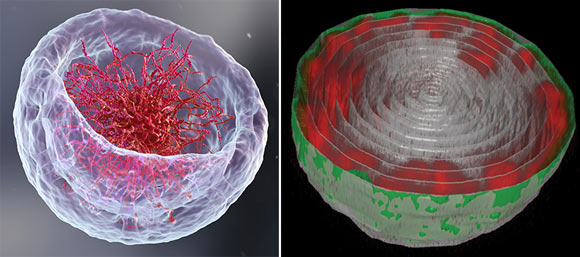
Left: a 3D illustration of the nucleus representing the classical theory of DNA organization at its center. Right: microscopic image of the nucleus of a fruit fly larva’s muscle cell; the long chains of DNA (red) are attached to the nuclear lamina (green) — the inner layer of the nuclear membrane. Image credit: Weizmann Institute of Science.
In an earlier study, Dr. Daria Amiad-Pavlov and her colleagues from the Weizmann Institute of Science analyzed how mechanical forces influence cell nuclei in the muscle and found evidence that muscle contractions had an immediate effect on gene expression patterns.
“We couldn’t explore this further because existing methods relied on imaging of chemically preserved cells, so they failed to capture what happens in the cell nuclei of an actual working muscle,” Dr. Amiad-Pavlov said.
To address this issue, the researchers aimed to study muscle nuclei in live fruit fly (Drosophila) larvae.
They obtained images of the internal, linearly-organized complexes of DNA and its proteins, surrounded by the membrane of the muscle nuclei.
Rather than filling up the entire volume of the nucleus, the ‘noodles,’ or long chromatin molecules, were organized as a relatively thin layer, attached to its inner walls.
Similar to the outcome of the interaction between oil and water, what is known as phase separation, the chromatin separated itself from the bulk of the liquid inside of the nucleus and found its place at its outskirts, while most of the fluid medium remained at the center.
The scientists realized that they were on their way to addressing a fundamental biological question, that is — how is chromatin, and hence DNA, organized in the nucleus in a living organism.
“But the findings were so unexpected, we had to make sure no error had crept in and that this organization was universal,” said Dr. Dana Lorber, also from the Weizmann Institute of Science.
The authors also built a theoretical model that included the physical factors governing chromatin organization in the nucleus, such as the relative forces of attraction between chromatin and its liquid environment and between chromatin and the nuclear membrane.
Their model predicted that the chromatin should undergo separation from the liquid phase, depending on the relative amount of liquid (hydration) in the nucleus. Furthermore, the phase separated chromatin could then arrange itself along the inside of the nuclear membrane — just as they had found in their experiments.
They also explained why chromatin appeared to fill the cell nuclei in previous studies.
“When scientists plate cells on a glass slide in order to study them under a microscope, they change their volume and physically flatten them,” said Weizmann Institute of Science’s Professor Samuel Safran.
“This may perturb some of the forces governing chromatin arrangement and reduce the distance between the upper part of the nucleus to its base.”
The team’s findings were published in the journal Science Advances.
_____
Daria Amiad-Pavlov et al. 2021. Live imaging of chromatin distribution reveals novel principles of nuclear architecture and chromatin compartmentalization. Science Advances 7 (23); doi: 10.1126/sciadv.abf6251







