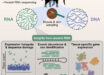
Scientists in Japan have generated a highly contiguous genome assembly of the red variety of Perilla frutescens, an important herbal plant with hundreds of bioactive chemicals, among which perillaldehyde and rosmarinic acid are the two major bioactive compounds in the plant.
Perilla frutescens is an annual herbal plant belonging to the family Lamiaceae, and it is widely cultivated in Asian countries.
There are two types of perilla plants based on the content of compounds called anthocyanins: red and green perilla.
Red perilla (‘aka-shiso’ in Japanese) is an anthocyanin-rich variety with dark red or purple leaves and stems, while green perilla (‘ao-jiso’ in Japanese) is an anthocyanin deficient variety with green leaves and stems.
Both green and red perilla leaves are often used as a material for cooking.
Particularly, the leaves of red perilla are used as traditional Kampo medicine ‘soyou’ to treat stomach problems.
The seeds are also used to produce oil, which is a rich source of alpha-linolenic acid.
The medicinal and nutritional uses of the red perilla could be improved by enhancing the production of valuable metabolites through the manipulation of key enzymes or regulatory genes using genome editing technology.
“Genome editing of red perilla for providing better traits is one of the promising ways to utilize this plant more effectively,” said Hiroshima University’s Professor Hidemasa Bono.
“To do so, high-quality genome sequences of the target species are necessary.”
For their research, ofessor Bono and colleagues collected the genetic material from young hydroponically grown leaves of Hoko-3, a specific cultivar of Perilla frutescens.
Their data show that Hoko-3 is highly genetically similar from plant to plant, in part because it is a self-fertilizing crop, making it ideal as a candidate for targeted gene editing.
“We achieved a highly contiguous genome assembly of red perilla using PacBio HiFi reads,” said Dr. Keita Tamura, also from Hiroshima University.
“We anchored 99.2% of the assembly into 20 pseudochromosomes, among which seven pseudochromosomes consisted of one contiguous segment.”
“Using systematic functional annotation workflow developed for plants called Fanflow4Plants, we functionally annotated 72,983 genes out of 76,825 protein coding genes,”
To annotate genes in the genome assembly, the authors used two complementary processes, each with unique strengths in sequencing.
“The number of gene models annotated in this study by combining two evidence-based annotations and the gene prediction method (BRAKER2) was 86,258, which is almost twice the previously assembled Perilla frutescens genome (43,527 genes) and close to the number of genes reported in another Lamiaceae tetraploid species Salvia splendens (88,489 genes),” they said.
“The genome assembly and functional annotations obtained in this study will be used for mining the target genes for genome editing of red perilla,” Dr. Tamura said.
“It could be possible to enhance the accumulation of valuable phytochemicals inside the plant.”
The results were published online in the journal DNA Research.
_____
Keita Tamura et al. A highly contiguous genome assembly of red perilla (Perilla frutescens) domesticated in Japan. DNA Research, published online November 16, 2022; doi: 10.1093/dnares/dsac044